You are viewing a plain text version of this content. The canonical link for it is here.
Posted to commits@spark.apache.org by ru...@apache.org on 2020/03/04 04:21:00 UTC
[spark] branch master updated: [SPARK-29212][ML][PYSPARK] Add
common classes without using JVM backend
This is an automated email from the ASF dual-hosted git repository.
ruifengz pushed a commit to branch master
in repository https://gitbox.apache.org/repos/asf/spark.git
The following commit(s) were added to refs/heads/master by this push:
new e1b3e9a [SPARK-29212][ML][PYSPARK] Add common classes without using JVM backend
e1b3e9a is described below
commit e1b3e9a3d25978dc0ad4609ecbc157ea1eebe2dd
Author: zero323 <ms...@gmail.com>
AuthorDate: Wed Mar 4 12:20:02 2020 +0800
[SPARK-29212][ML][PYSPARK] Add common classes without using JVM backend
### What changes were proposed in this pull request?
Implement common base ML classes (`Predictor`, `PredictionModel`, `Classifier`, `ClasssificationModel` `ProbabilisticClassifier`, `ProbabilisticClasssificationModel`, `Regressor`, `RegrssionModel`) for non-Java backends.
Note
- `Predictor` and `JavaClassifier` should be abstract as `_fit` method is not implemented.
- `PredictionModel` should be abstract as `_transform` is not implemented.
### Why are the changes needed?
To provide extensions points for non-JVM algorithms, as well as a public (as opposed to `Java*` variants, which are commonly described in docstrings as private) hierarchy which can be used to distinguish between different classes of predictors.
For longer discussion see [SPARK-29212](https://issues.apache.org/jira/browse/SPARK-29212) and / or https://github.com/apache/spark/pull/25776.
### Does this PR introduce any user-facing change?
It adds new base classes as listed above, but effective interfaces (method resolution order notwithstanding) stay the same.
Additionally "private" `Java*` classes in`ml.regression` and `ml.classification` have been renamed to follow PEP-8 conventions (added leading underscore).
It is for discussion if the same should be done to equivalent classes from `ml.wrapper`.
If we take `JavaClassifier` as an example, type hierarchy will change from
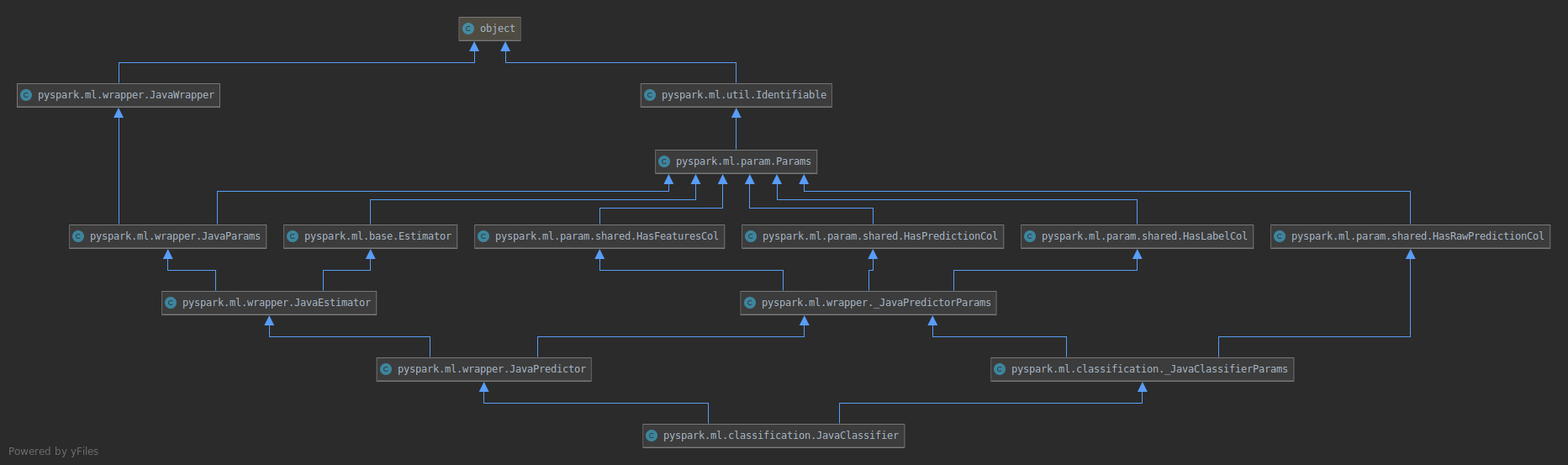
to
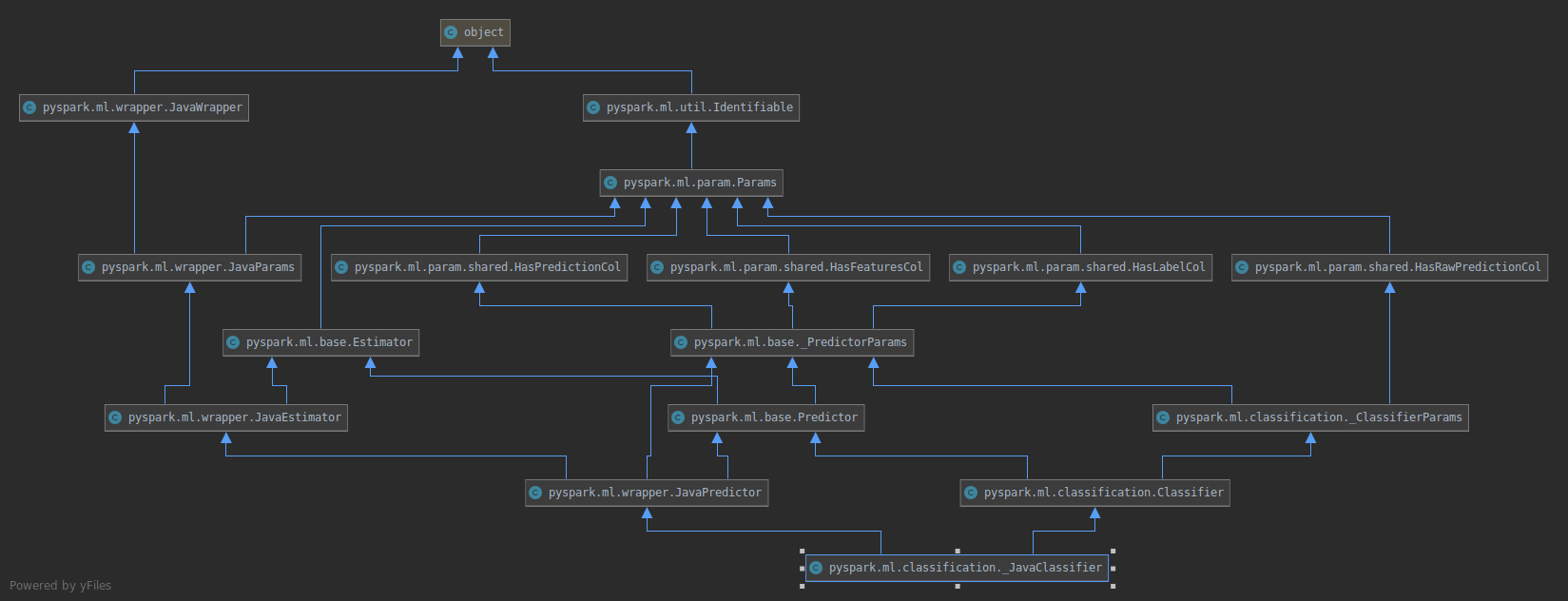
Similarly the old model
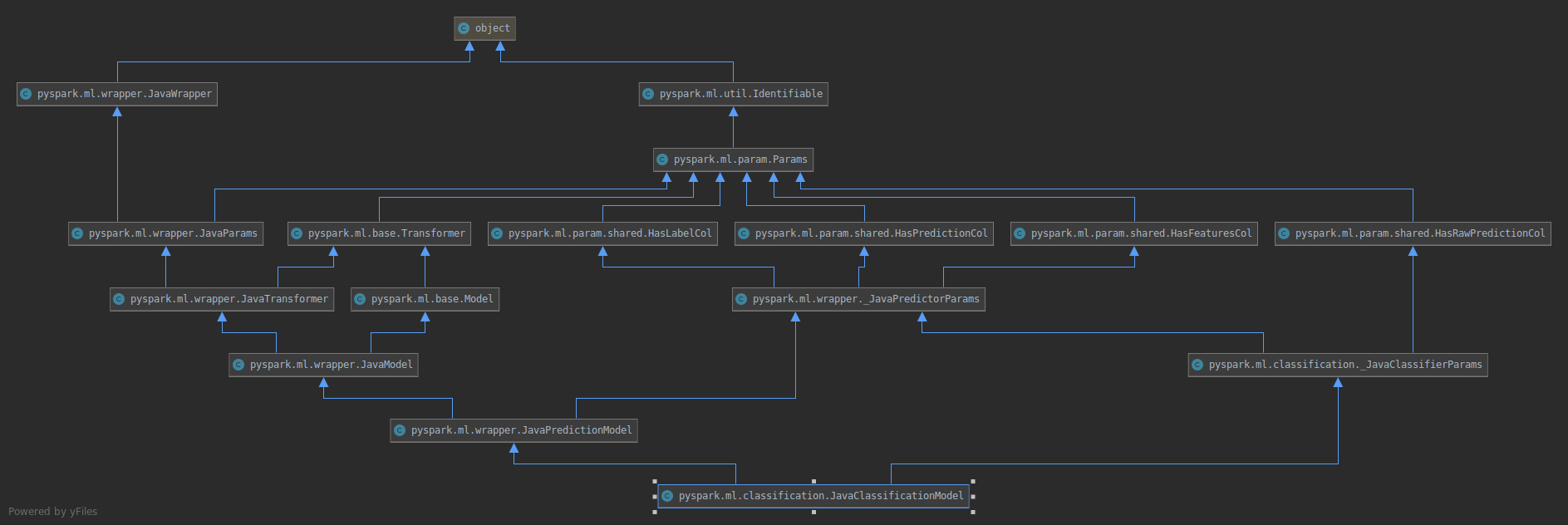
will become
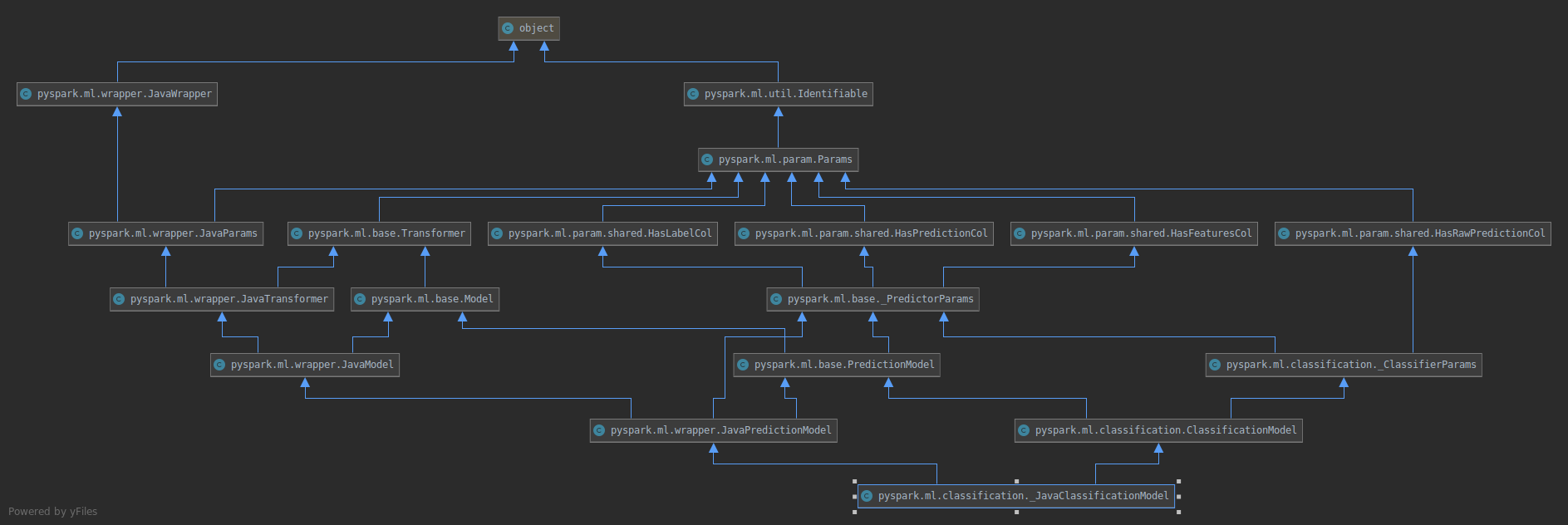
### How was this patch tested?
Existing unit tests.
Closes #27245 from zero323/SPARK-29212.
Authored-by: zero323 <ms...@gmail.com>
Signed-off-by: zhengruifeng <ru...@foxmail.com>
---
python/pyspark/ml/__init__.py | 6 +-
python/pyspark/ml/base.py | 81 ++++++++++++++++-
python/pyspark/ml/classification.py | 158 +++++++++++++++++++++++++---------
python/pyspark/ml/regression.py | 71 ++++++++++-----
python/pyspark/ml/tests/test_param.py | 6 +-
python/pyspark/ml/wrapper.py | 52 ++---------
6 files changed, 258 insertions(+), 116 deletions(-)
diff --git a/python/pyspark/ml/__init__.py b/python/pyspark/ml/__init__.py
index d99a253..47fc78e 100644
--- a/python/pyspark/ml/__init__.py
+++ b/python/pyspark/ml/__init__.py
@@ -19,13 +19,15 @@
DataFrame-based machine learning APIs to let users quickly assemble and configure practical
machine learning pipelines.
"""
-from pyspark.ml.base import Estimator, Model, Transformer, UnaryTransformer
+from pyspark.ml.base import Estimator, Model, Predictor, PredictionModel, \
+ Transformer, UnaryTransformer
from pyspark.ml.pipeline import Pipeline, PipelineModel
from pyspark.ml import classification, clustering, evaluation, feature, fpm, \
image, pipeline, recommendation, regression, stat, tuning, util, linalg, param
__all__ = [
- "Transformer", "UnaryTransformer", "Estimator", "Model", "Pipeline", "PipelineModel",
+ "Transformer", "UnaryTransformer", "Estimator", "Model",
+ "Predictor", "PredictionModel", "Pipeline", "PipelineModel",
"classification", "clustering", "evaluation", "feature", "fpm", "image",
"recommendation", "regression", "stat", "tuning", "util", "linalg", "param",
]
diff --git a/python/pyspark/ml/base.py b/python/pyspark/ml/base.py
index 542cb25..b8df5a3 100644
--- a/python/pyspark/ml/base.py
+++ b/python/pyspark/ml/base.py
@@ -15,7 +15,7 @@
# limitations under the License.
#
-from abc import ABCMeta, abstractmethod
+from abc import ABCMeta, abstractmethod, abstractproperty
import copy
import threading
@@ -246,3 +246,82 @@ class UnaryTransformer(HasInputCol, HasOutputCol, Transformer):
transformedDataset = dataset.withColumn(self.getOutputCol(),
transformUDF(dataset[self.getInputCol()]))
return transformedDataset
+
+
+@inherit_doc
+class _PredictorParams(HasLabelCol, HasFeaturesCol, HasPredictionCol):
+ """
+ Params for :py:class:`Predictor` and :py:class:`PredictorModel`.
+
+ .. versionadded:: 3.0.0
+ """
+ pass
+
+
+@inherit_doc
+class Predictor(Estimator, _PredictorParams):
+ """
+ Estimator for prediction tasks (regression and classification).
+ """
+
+ __metaclass__ = ABCMeta
+
+ @since("3.0.0")
+ def setLabelCol(self, value):
+ """
+ Sets the value of :py:attr:`labelCol`.
+ """
+ return self._set(labelCol=value)
+
+ @since("3.0.0")
+ def setFeaturesCol(self, value):
+ """
+ Sets the value of :py:attr:`featuresCol`.
+ """
+ return self._set(featuresCol=value)
+
+ @since("3.0.0")
+ def setPredictionCol(self, value):
+ """
+ Sets the value of :py:attr:`predictionCol`.
+ """
+ return self._set(predictionCol=value)
+
+
+@inherit_doc
+class PredictionModel(Model, _PredictorParams):
+ """
+ Model for prediction tasks (regression and classification).
+ """
+
+ __metaclass__ = ABCMeta
+
+ @since("3.0.0")
+ def setFeaturesCol(self, value):
+ """
+ Sets the value of :py:attr:`featuresCol`.
+ """
+ return self._set(featuresCol=value)
+
+ @since("3.0.0")
+ def setPredictionCol(self, value):
+ """
+ Sets the value of :py:attr:`predictionCol`.
+ """
+ return self._set(predictionCol=value)
+
+ @abstractproperty
+ @since("2.1.0")
+ def numFeatures(self):
+ """
+ Returns the number of features the model was trained on. If unknown, returns -1
+ """
+ raise NotImplementedError()
+
+ @abstractmethod
+ @since("3.0.0")
+ def predict(self, value):
+ """
+ Predict label for the given features.
+ """
+ raise NotImplementedError()
diff --git a/python/pyspark/ml/classification.py b/python/pyspark/ml/classification.py
index 1436b78..0d88aa8 100644
--- a/python/pyspark/ml/classification.py
+++ b/python/pyspark/ml/classification.py
@@ -17,18 +17,20 @@
import operator
import sys
+from abc import ABCMeta, abstractmethod, abstractproperty
from multiprocessing.pool import ThreadPool
from pyspark import since, keyword_only
-from pyspark.ml import Estimator, Model
+from pyspark.ml import Estimator, Predictor, PredictionModel, Model
from pyspark.ml.param.shared import *
from pyspark.ml.tree import _DecisionTreeModel, _DecisionTreeParams, \
_TreeEnsembleModel, _RandomForestParams, _GBTParams, \
_HasVarianceImpurity, _TreeClassifierParams, _TreeEnsembleParams
from pyspark.ml.regression import _FactorizationMachinesParams, DecisionTreeRegressionModel
from pyspark.ml.util import *
+from pyspark.ml.base import _PredictorParams
from pyspark.ml.wrapper import JavaEstimator, JavaModel, JavaParams, \
- JavaPredictor, _JavaPredictorParams, JavaPredictionModel, JavaWrapper
+ JavaPredictor, JavaPredictionModel, JavaWrapper
from pyspark.ml.common import inherit_doc, _java2py, _py2java
from pyspark.ml.linalg import Vectors
from pyspark.sql import DataFrame
@@ -49,9 +51,9 @@ __all__ = ['LinearSVC', 'LinearSVCModel',
'FMClassifier', 'FMClassificationModel']
-class _JavaClassifierParams(HasRawPredictionCol, _JavaPredictorParams):
+class _ClassifierParams(HasRawPredictionCol, _PredictorParams):
"""
- Java Classifier Params for classification tasks.
+ Classifier Params for classification tasks.
.. versionadded:: 3.0.0
"""
@@ -59,12 +61,14 @@ class _JavaClassifierParams(HasRawPredictionCol, _JavaPredictorParams):
@inherit_doc
-class JavaClassifier(JavaPredictor, _JavaClassifierParams):
+class Classifier(Predictor, _ClassifierParams):
"""
- Java Classifier for classification tasks.
+ Classifier for classification tasks.
Classes are indexed {0, 1, ..., numClasses - 1}.
"""
+ __metaclass__ = ABCMeta
+
@since("3.0.0")
def setRawPredictionCol(self, value):
"""
@@ -74,13 +78,14 @@ class JavaClassifier(JavaPredictor, _JavaClassifierParams):
@inherit_doc
-class JavaClassificationModel(JavaPredictionModel, _JavaClassifierParams):
+class ClassificationModel(PredictionModel, _ClassifierParams):
"""
- Java Model produced by a ``Classifier``.
+ Model produced by a ``Classifier``.
Classes are indexed {0, 1, ..., numClasses - 1}.
- To be mixed in with class:`pyspark.ml.JavaModel`
"""
+ __metaclass__ = ABCMeta
+
@since("3.0.0")
def setRawPredictionCol(self, value):
"""
@@ -88,26 +93,27 @@ class JavaClassificationModel(JavaPredictionModel, _JavaClassifierParams):
"""
return self._set(rawPredictionCol=value)
- @property
+ @abstractproperty
@since("2.1.0")
def numClasses(self):
"""
Number of classes (values which the label can take).
"""
- return self._call_java("numClasses")
+ raise NotImplementedError()
+ @abstractmethod
@since("3.0.0")
def predictRaw(self, value):
"""
Raw prediction for each possible label.
"""
- return self._call_java("predictRaw", value)
+ raise NotImplementedError()
-class _JavaProbabilisticClassifierParams(HasProbabilityCol, HasThresholds, _JavaClassifierParams):
+class _ProbabilisticClassifierParams(HasProbabilityCol, HasThresholds, _ClassifierParams):
"""
- Params for :py:class:`JavaProbabilisticClassifier` and
- :py:class:`JavaProbabilisticClassificationModel`.
+ Params for :py:class:`ProbabilisticClassifier` and
+ :py:class:`ProbabilisticClassificationModel`.
.. versionadded:: 3.0.0
"""
@@ -115,11 +121,13 @@ class _JavaProbabilisticClassifierParams(HasProbabilityCol, HasThresholds, _Java
@inherit_doc
-class JavaProbabilisticClassifier(JavaClassifier, _JavaProbabilisticClassifierParams):
+class ProbabilisticClassifier(Classifier, _ProbabilisticClassifierParams):
"""
- Java Probabilistic Classifier for classification tasks.
+ Probabilistic Classifier for classification tasks.
"""
+ __metaclass__ = ABCMeta
+
@since("3.0.0")
def setProbabilityCol(self, value):
"""
@@ -136,12 +144,14 @@ class JavaProbabilisticClassifier(JavaClassifier, _JavaProbabilisticClassifierPa
@inherit_doc
-class JavaProbabilisticClassificationModel(JavaClassificationModel,
- _JavaProbabilisticClassifierParams):
+class ProbabilisticClassificationModel(ClassificationModel,
+ _ProbabilisticClassifierParams):
"""
- Java Model produced by a ``ProbabilisticClassifier``.
+ Model produced by a ``ProbabilisticClassifier``.
"""
+ __metaclass__ = ABCMeta
+
@since("3.0.0")
def setProbabilityCol(self, value):
"""
@@ -156,6 +166,72 @@ class JavaProbabilisticClassificationModel(JavaClassificationModel,
"""
return self._set(thresholds=value)
+ @abstractmethod
+ @since("3.0.0")
+ def predictProbability(self, value):
+ """
+ Predict the probability of each class given the features.
+ """
+ raise NotImplementedError()
+
+
+@inherit_doc
+class _JavaClassifier(Classifier, JavaPredictor):
+ """
+ Java Classifier for classification tasks.
+ Classes are indexed {0, 1, ..., numClasses - 1}.
+ """
+
+ __metaclass__ = ABCMeta
+
+ @since("3.0.0")
+ def setRawPredictionCol(self, value):
+ """
+ Sets the value of :py:attr:`rawPredictionCol`.
+ """
+ return self._set(rawPredictionCol=value)
+
+
+@inherit_doc
+class _JavaClassificationModel(ClassificationModel, JavaPredictionModel):
+ """
+ Java Model produced by a ``Classifier``.
+ Classes are indexed {0, 1, ..., numClasses - 1}.
+ To be mixed in with class:`pyspark.ml.JavaModel`
+ """
+
+ @property
+ @since("2.1.0")
+ def numClasses(self):
+ """
+ Number of classes (values which the label can take).
+ """
+ return self._call_java("numClasses")
+
+ @since("3.0.0")
+ def predictRaw(self, value):
+ """
+ Raw prediction for each possible label.
+ """
+ return self._call_java("predictRaw", value)
+
+
+@inherit_doc
+class _JavaProbabilisticClassifier(ProbabilisticClassifier, _JavaClassifier):
+ """
+ Java Probabilistic Classifier for classification tasks.
+ """
+
+ __metaclass__ = ABCMeta
+
+
+@inherit_doc
+class _JavaProbabilisticClassificationModel(ProbabilisticClassificationModel,
+ _JavaClassificationModel):
+ """
+ Java Model produced by a ``ProbabilisticClassifier``.
+ """
+
@since("3.0.0")
def predictProbability(self, value):
"""
@@ -164,7 +240,7 @@ class JavaProbabilisticClassificationModel(JavaClassificationModel,
return self._call_java("predictProbability", value)
-class _LinearSVCParams(_JavaClassifierParams, HasRegParam, HasMaxIter, HasFitIntercept, HasTol,
+class _LinearSVCParams(_ClassifierParams, HasRegParam, HasMaxIter, HasFitIntercept, HasTol,
HasStandardization, HasWeightCol, HasAggregationDepth, HasThreshold):
"""
Params for :py:class:`LinearSVC` and :py:class:`LinearSVCModel`.
@@ -180,7 +256,7 @@ class _LinearSVCParams(_JavaClassifierParams, HasRegParam, HasMaxIter, HasFitInt
@inherit_doc
-class LinearSVC(JavaClassifier, _LinearSVCParams, JavaMLWritable, JavaMLReadable):
+class LinearSVC(_JavaClassifier, _LinearSVCParams, JavaMLWritable, JavaMLReadable):
"""
`Linear SVM Classifier <https://en.wikipedia.org/wiki/Support_vector_machine#Linear_SVM>`_
@@ -343,7 +419,7 @@ class LinearSVC(JavaClassifier, _LinearSVCParams, JavaMLWritable, JavaMLReadable
return self._set(aggregationDepth=value)
-class LinearSVCModel(JavaClassificationModel, _LinearSVCParams, JavaMLWritable, JavaMLReadable):
+class LinearSVCModel(_JavaClassificationModel, _LinearSVCParams, JavaMLWritable, JavaMLReadable):
"""
Model fitted by LinearSVC.
@@ -374,7 +450,7 @@ class LinearSVCModel(JavaClassificationModel, _LinearSVCParams, JavaMLWritable,
return self._call_java("intercept")
-class _LogisticRegressionParams(_JavaProbabilisticClassifierParams, HasRegParam,
+class _LogisticRegressionParams(_ProbabilisticClassifierParams, HasRegParam,
HasElasticNetParam, HasMaxIter, HasFitIntercept, HasTol,
HasStandardization, HasWeightCol, HasAggregationDepth,
HasThreshold):
@@ -533,7 +609,7 @@ class _LogisticRegressionParams(_JavaProbabilisticClassifierParams, HasRegParam,
@inherit_doc
-class LogisticRegression(JavaProbabilisticClassifier, _LogisticRegressionParams, JavaMLWritable,
+class LogisticRegression(_JavaProbabilisticClassifier, _LogisticRegressionParams, JavaMLWritable,
JavaMLReadable):
"""
Logistic regression.
@@ -759,7 +835,7 @@ class LogisticRegression(JavaProbabilisticClassifier, _LogisticRegressionParams,
return self._set(aggregationDepth=value)
-class LogisticRegressionModel(JavaProbabilisticClassificationModel, _LogisticRegressionParams,
+class LogisticRegressionModel(_JavaProbabilisticClassificationModel, _LogisticRegressionParams,
JavaMLWritable, JavaMLReadable, HasTrainingSummary):
"""
Model fitted by LogisticRegression.
@@ -1131,7 +1207,7 @@ class _DecisionTreeClassifierParams(_DecisionTreeParams, _TreeClassifierParams):
@inherit_doc
-class DecisionTreeClassifier(JavaProbabilisticClassifier, _DecisionTreeClassifierParams,
+class DecisionTreeClassifier(_JavaProbabilisticClassifier, _DecisionTreeClassifierParams,
JavaMLWritable, JavaMLReadable):
"""
`Decision tree <http://en.wikipedia.org/wiki/Decision_tree_learning>`_
@@ -1326,7 +1402,7 @@ class DecisionTreeClassifier(JavaProbabilisticClassifier, _DecisionTreeClassifie
@inherit_doc
-class DecisionTreeClassificationModel(_DecisionTreeModel, JavaProbabilisticClassificationModel,
+class DecisionTreeClassificationModel(_DecisionTreeModel, _JavaProbabilisticClassificationModel,
_DecisionTreeClassifierParams, JavaMLWritable,
JavaMLReadable):
"""
@@ -1366,7 +1442,7 @@ class _RandomForestClassifierParams(_RandomForestParams, _TreeClassifierParams):
@inherit_doc
-class RandomForestClassifier(JavaProbabilisticClassifier, _RandomForestClassifierParams,
+class RandomForestClassifier(_JavaProbabilisticClassifier, _RandomForestClassifierParams,
JavaMLWritable, JavaMLReadable):
"""
`Random Forest <http://en.wikipedia.org/wiki/Random_forest>`_
@@ -1585,7 +1661,7 @@ class RandomForestClassifier(JavaProbabilisticClassifier, _RandomForestClassifie
return self._set(minWeightFractionPerNode=value)
-class RandomForestClassificationModel(_TreeEnsembleModel, JavaProbabilisticClassificationModel,
+class RandomForestClassificationModel(_TreeEnsembleModel, _JavaProbabilisticClassificationModel,
_RandomForestClassifierParams, JavaMLWritable,
JavaMLReadable):
"""
@@ -1639,7 +1715,7 @@ class _GBTClassifierParams(_GBTParams, _HasVarianceImpurity):
@inherit_doc
-class GBTClassifier(JavaProbabilisticClassifier, _GBTClassifierParams,
+class GBTClassifier(_JavaProbabilisticClassifier, _GBTClassifierParams,
JavaMLWritable, JavaMLReadable):
"""
`Gradient-Boosted Trees (GBTs) <http://en.wikipedia.org/wiki/Gradient_boosting>`_
@@ -1904,7 +1980,7 @@ class GBTClassifier(JavaProbabilisticClassifier, _GBTClassifierParams,
return self._set(minWeightFractionPerNode=value)
-class GBTClassificationModel(_TreeEnsembleModel, JavaProbabilisticClassificationModel,
+class GBTClassificationModel(_TreeEnsembleModel, _JavaProbabilisticClassificationModel,
_GBTClassifierParams, JavaMLWritable, JavaMLReadable):
"""
Model fitted by GBTClassifier.
@@ -1945,7 +2021,7 @@ class GBTClassificationModel(_TreeEnsembleModel, JavaProbabilisticClassification
return self._call_java("evaluateEachIteration", dataset)
-class _NaiveBayesParams(_JavaPredictorParams, HasWeightCol):
+class _NaiveBayesParams(_PredictorParams, HasWeightCol):
"""
Params for :py:class:`NaiveBayes` and :py:class:`NaiveBayesModel`.
@@ -1975,7 +2051,7 @@ class _NaiveBayesParams(_JavaPredictorParams, HasWeightCol):
@inherit_doc
-class NaiveBayes(JavaProbabilisticClassifier, _NaiveBayesParams, HasThresholds, HasWeightCol,
+class NaiveBayes(_JavaProbabilisticClassifier, _NaiveBayesParams, HasThresholds, HasWeightCol,
JavaMLWritable, JavaMLReadable):
"""
Naive Bayes Classifiers.
@@ -2119,7 +2195,7 @@ class NaiveBayes(JavaProbabilisticClassifier, _NaiveBayesParams, HasThresholds,
return self._set(weightCol=value)
-class NaiveBayesModel(JavaProbabilisticClassificationModel, _NaiveBayesParams, JavaMLWritable,
+class NaiveBayesModel(_JavaProbabilisticClassificationModel, _NaiveBayesParams, JavaMLWritable,
JavaMLReadable):
"""
Model fitted by NaiveBayes.
@@ -2152,7 +2228,7 @@ class NaiveBayesModel(JavaProbabilisticClassificationModel, _NaiveBayesParams, J
return self._call_java("sigma")
-class _MultilayerPerceptronParams(_JavaProbabilisticClassifierParams, HasSeed, HasMaxIter,
+class _MultilayerPerceptronParams(_ProbabilisticClassifierParams, HasSeed, HasMaxIter,
HasTol, HasStepSize, HasSolver, HasBlockSize):
"""
Params for :py:class:`MultilayerPerceptronClassifier`.
@@ -2185,7 +2261,7 @@ class _MultilayerPerceptronParams(_JavaProbabilisticClassifierParams, HasSeed, H
@inherit_doc
-class MultilayerPerceptronClassifier(JavaProbabilisticClassifier, _MultilayerPerceptronParams,
+class MultilayerPerceptronClassifier(_JavaProbabilisticClassifier, _MultilayerPerceptronParams,
JavaMLWritable, JavaMLReadable):
"""
Classifier trainer based on the Multilayer Perceptron.
@@ -2348,7 +2424,7 @@ class MultilayerPerceptronClassifier(JavaProbabilisticClassifier, _MultilayerPer
return self._set(solver=value)
-class MultilayerPerceptronClassificationModel(JavaProbabilisticClassificationModel,
+class MultilayerPerceptronClassificationModel(_JavaProbabilisticClassificationModel,
_MultilayerPerceptronParams, JavaMLWritable,
JavaMLReadable):
"""
@@ -2366,7 +2442,7 @@ class MultilayerPerceptronClassificationModel(JavaProbabilisticClassificationMod
return self._call_java("weights")
-class _OneVsRestParams(_JavaClassifierParams, HasWeightCol):
+class _OneVsRestParams(_ClassifierParams, HasWeightCol):
"""
Params for :py:class:`OneVsRest` and :py:class:`OneVsRestModelModel`.
"""
@@ -2802,7 +2878,7 @@ class OneVsRestModel(Model, _OneVsRestParams, JavaMLReadable, JavaMLWritable):
@inherit_doc
-class FMClassifier(JavaProbabilisticClassifier, _FactorizationMachinesParams, JavaMLWritable,
+class FMClassifier(_JavaProbabilisticClassifier, _FactorizationMachinesParams, JavaMLWritable,
JavaMLReadable):
"""
Factorization Machines learning algorithm for classification.
@@ -2973,7 +3049,7 @@ class FMClassifier(JavaProbabilisticClassifier, _FactorizationMachinesParams, Ja
return self._set(regParam=value)
-class FMClassificationModel(JavaProbabilisticClassificationModel, _FactorizationMachinesParams,
+class FMClassificationModel(_JavaProbabilisticClassificationModel, _FactorizationMachinesParams,
JavaMLWritable, JavaMLReadable):
"""
Model fitted by :class:`FMClassifier`.
diff --git a/python/pyspark/ml/regression.py b/python/pyspark/ml/regression.py
index a4c9782..f227fe0 100644
--- a/python/pyspark/ml/regression.py
+++ b/python/pyspark/ml/regression.py
@@ -16,15 +16,18 @@
#
import sys
+from abc import ABCMeta
from pyspark import since, keyword_only
+from pyspark.ml import Predictor, PredictionModel
+from pyspark.ml.base import _PredictorParams
from pyspark.ml.param.shared import *
from pyspark.ml.tree import _DecisionTreeModel, _DecisionTreeParams, \
_TreeEnsembleModel, _TreeEnsembleParams, _RandomForestParams, _GBTParams, \
_HasVarianceImpurity, _TreeRegressorParams
from pyspark.ml.util import *
from pyspark.ml.wrapper import JavaEstimator, JavaModel, JavaParams, \
- JavaPredictor, JavaPredictionModel, _JavaPredictorParams, JavaWrapper
+ JavaPredictor, JavaPredictionModel, JavaWrapper
from pyspark.ml.common import inherit_doc
from pyspark.sql import DataFrame
@@ -41,26 +44,48 @@ __all__ = ['AFTSurvivalRegression', 'AFTSurvivalRegressionModel',
'FMRegressor', 'FMRegressionModel']
-class JavaRegressor(JavaPredictor, _JavaPredictorParams):
+class Regressor(Predictor, _PredictorParams):
+ """
+ Regressor for regression tasks.
+
+ .. versionadded:: 3.0.0
+ """
+
+ __metaclass__ = ABCMeta
+
+
+class RegressionModel(PredictionModel, _PredictorParams):
+ """
+ Model produced by a ``Regressor``.
+
+ .. versionadded:: 3.0.0
+ """
+
+ __metaclass__ = ABCMeta
+
+
+class _JavaRegressor(Regressor, JavaPredictor):
"""
Java Regressor for regression tasks.
.. versionadded:: 3.0.0
"""
- pass
+
+ __metaclass__ = ABCMeta
-class JavaRegressionModel(JavaPredictionModel, _JavaPredictorParams):
+class _JavaRegressionModel(RegressionModel, JavaPredictionModel):
"""
Java Model produced by a ``_JavaRegressor``.
To be mixed in with class:`pyspark.ml.JavaModel`
.. versionadded:: 3.0.0
"""
- pass
+
+ __metaclass__ = ABCMeta
-class _LinearRegressionParams(_JavaPredictorParams, HasRegParam, HasElasticNetParam, HasMaxIter,
+class _LinearRegressionParams(_PredictorParams, HasRegParam, HasElasticNetParam, HasMaxIter,
HasTol, HasFitIntercept, HasStandardization, HasWeightCol, HasSolver,
HasAggregationDepth, HasLoss):
"""
@@ -88,7 +113,7 @@ class _LinearRegressionParams(_JavaPredictorParams, HasRegParam, HasElasticNetPa
@inherit_doc
-class LinearRegression(JavaRegressor, _LinearRegressionParams, JavaMLWritable, JavaMLReadable):
+class LinearRegression(_JavaRegressor, _LinearRegressionParams, JavaMLWritable, JavaMLReadable):
"""
Linear regression.
@@ -270,7 +295,7 @@ class LinearRegression(JavaRegressor, _LinearRegressionParams, JavaMLWritable, J
return self._set(lossType=value)
-class LinearRegressionModel(JavaRegressionModel, _LinearRegressionParams, GeneralJavaMLWritable,
+class LinearRegressionModel(_JavaRegressionModel, _LinearRegressionParams, GeneralJavaMLWritable,
JavaMLReadable, HasTrainingSummary):
"""
Model fitted by :class:`LinearRegression`.
@@ -777,7 +802,7 @@ class _DecisionTreeRegressorParams(_DecisionTreeParams, _TreeRegressorParams, Ha
@inherit_doc
-class DecisionTreeRegressor(JavaRegressor, _DecisionTreeRegressorParams, JavaMLWritable,
+class DecisionTreeRegressor(_JavaRegressor, _DecisionTreeRegressorParams, JavaMLWritable,
JavaMLReadable):
"""
`Decision tree <http://en.wikipedia.org/wiki/Decision_tree_learning>`_
@@ -973,7 +998,7 @@ class DecisionTreeRegressor(JavaRegressor, _DecisionTreeRegressorParams, JavaMLW
@inherit_doc
class DecisionTreeRegressionModel(
- JavaRegressionModel, _DecisionTreeModel, _DecisionTreeRegressorParams,
+ _JavaRegressionModel, _DecisionTreeModel, _DecisionTreeRegressorParams,
JavaMLWritable, JavaMLReadable
):
"""
@@ -1021,7 +1046,7 @@ class _RandomForestRegressorParams(_RandomForestParams, _TreeRegressorParams):
@inherit_doc
-class RandomForestRegressor(JavaRegressor, _RandomForestRegressorParams, JavaMLWritable,
+class RandomForestRegressor(_JavaRegressor, _RandomForestRegressorParams, JavaMLWritable,
JavaMLReadable):
"""
`Random Forest <http://en.wikipedia.org/wiki/Random_forest>`_
@@ -1230,7 +1255,7 @@ class RandomForestRegressor(JavaRegressor, _RandomForestRegressorParams, JavaMLW
class RandomForestRegressionModel(
- JavaRegressionModel, _TreeEnsembleModel, _RandomForestRegressorParams,
+ _JavaRegressionModel, _TreeEnsembleModel, _RandomForestRegressorParams,
JavaMLWritable, JavaMLReadable
):
"""
@@ -1284,7 +1309,7 @@ class _GBTRegressorParams(_GBTParams, _TreeRegressorParams):
@inherit_doc
-class GBTRegressor(JavaRegressor, _GBTRegressorParams, JavaMLWritable, JavaMLReadable):
+class GBTRegressor(_JavaRegressor, _GBTRegressorParams, JavaMLWritable, JavaMLReadable):
"""
`Gradient-Boosted Trees (GBTs) <http://en.wikipedia.org/wiki/Gradient_boosting>`_
learning algorithm for regression.
@@ -1526,7 +1551,7 @@ class GBTRegressor(JavaRegressor, _GBTRegressorParams, JavaMLWritable, JavaMLRea
class GBTRegressionModel(
- JavaRegressionModel, _TreeEnsembleModel, _GBTRegressorParams,
+ _JavaRegressionModel, _TreeEnsembleModel, _GBTRegressorParams,
JavaMLWritable, JavaMLReadable
):
"""
@@ -1571,7 +1596,7 @@ class GBTRegressionModel(
return self._call_java("evaluateEachIteration", dataset, loss)
-class _AFTSurvivalRegressionParams(_JavaPredictorParams, HasMaxIter, HasTol, HasFitIntercept,
+class _AFTSurvivalRegressionParams(_PredictorParams, HasMaxIter, HasTol, HasFitIntercept,
HasAggregationDepth):
"""
Params for :py:class:`AFTSurvivalRegression` and :py:class:`AFTSurvivalRegressionModel`.
@@ -1618,7 +1643,7 @@ class _AFTSurvivalRegressionParams(_JavaPredictorParams, HasMaxIter, HasTol, Has
@inherit_doc
-class AFTSurvivalRegression(JavaRegressor, _AFTSurvivalRegressionParams,
+class AFTSurvivalRegression(_JavaRegressor, _AFTSurvivalRegressionParams,
JavaMLWritable, JavaMLReadable):
"""
Accelerated Failure Time (AFT) Model Survival Regression
@@ -1759,7 +1784,7 @@ class AFTSurvivalRegression(JavaRegressor, _AFTSurvivalRegressionParams,
return self._set(aggregationDepth=value)
-class AFTSurvivalRegressionModel(JavaRegressionModel, _AFTSurvivalRegressionParams,
+class AFTSurvivalRegressionModel(_JavaRegressionModel, _AFTSurvivalRegressionParams,
JavaMLWritable, JavaMLReadable):
"""
Model fitted by :class:`AFTSurvivalRegression`.
@@ -1813,7 +1838,7 @@ class AFTSurvivalRegressionModel(JavaRegressionModel, _AFTSurvivalRegressionPara
return self._call_java("predictQuantiles", features)
-class _GeneralizedLinearRegressionParams(_JavaPredictorParams, HasFitIntercept, HasMaxIter,
+class _GeneralizedLinearRegressionParams(_PredictorParams, HasFitIntercept, HasMaxIter,
HasTol, HasRegParam, HasWeightCol, HasSolver,
HasAggregationDepth):
"""
@@ -1891,7 +1916,7 @@ class _GeneralizedLinearRegressionParams(_JavaPredictorParams, HasFitIntercept,
@inherit_doc
-class GeneralizedLinearRegression(JavaRegressor, _GeneralizedLinearRegressionParams,
+class GeneralizedLinearRegression(_JavaRegressor, _GeneralizedLinearRegressionParams,
JavaMLWritable, JavaMLReadable):
"""
Generalized Linear Regression.
@@ -2096,7 +2121,7 @@ class GeneralizedLinearRegression(JavaRegressor, _GeneralizedLinearRegressionPar
return self._set(aggregationDepth=value)
-class GeneralizedLinearRegressionModel(JavaRegressionModel, _GeneralizedLinearRegressionParams,
+class GeneralizedLinearRegressionModel(_JavaRegressionModel, _GeneralizedLinearRegressionParams,
JavaMLWritable, JavaMLReadable, HasTrainingSummary):
"""
Model fitted by :class:`GeneralizedLinearRegression`.
@@ -2328,7 +2353,7 @@ class GeneralizedLinearRegressionTrainingSummary(GeneralizedLinearRegressionSumm
return self._call_java("toString")
-class _FactorizationMachinesParams(_JavaPredictorParams, HasMaxIter, HasStepSize, HasTol,
+class _FactorizationMachinesParams(_PredictorParams, HasMaxIter, HasStepSize, HasTol,
HasSolver, HasSeed, HasFitIntercept, HasRegParam):
"""
Params for :py:class:`FMRegressor`, :py:class:`FMRegressionModel`, :py:class:`FMClassifier`
@@ -2384,7 +2409,7 @@ class _FactorizationMachinesParams(_JavaPredictorParams, HasMaxIter, HasStepSize
@inherit_doc
-class FMRegressor(JavaRegressor, _FactorizationMachinesParams, JavaMLWritable, JavaMLReadable):
+class FMRegressor(_JavaRegressor, _FactorizationMachinesParams, JavaMLWritable, JavaMLReadable):
"""
Factorization Machines learning algorithm for regression.
@@ -2548,7 +2573,7 @@ class FMRegressor(JavaRegressor, _FactorizationMachinesParams, JavaMLWritable, J
return self._set(regParam=value)
-class FMRegressionModel(JavaRegressionModel, _FactorizationMachinesParams, JavaMLWritable,
+class FMRegressionModel(_JavaRegressionModel, _FactorizationMachinesParams, JavaMLWritable,
JavaMLReadable):
"""
Model fitted by :class:`FMRegressor`.
diff --git a/python/pyspark/ml/tests/test_param.py b/python/pyspark/ml/tests/test_param.py
index 777b493..61f9f18 100644
--- a/python/pyspark/ml/tests/test_param.py
+++ b/python/pyspark/ml/tests/test_param.py
@@ -348,8 +348,9 @@ class DefaultValuesTests(PySparkTestCase):
Test :py:class:`JavaParams` classes to see if their default Param values match
those in their Scala counterparts.
"""
-
def test_java_params(self):
+ import re
+
import pyspark.ml.feature
import pyspark.ml.classification
import pyspark.ml.clustering
@@ -365,8 +366,9 @@ class DefaultValuesTests(PySparkTestCase):
for name, cls in inspect.getmembers(module, inspect.isclass):
if not name.endswith('Model') and not name.endswith('Params') \
and issubclass(cls, JavaParams) and not inspect.isabstract(cls) \
- and not name.startswith('Java') and name != '_LSH':
+ and not re.match("_?Java", name) and name != '_LSH':
# NOTE: disable check_params_exist until there is parity with Scala API
+
check_params(self, cls(), check_params_exist=False)
# Additional classes that need explicit construction
diff --git a/python/pyspark/ml/wrapper.py b/python/pyspark/ml/wrapper.py
index ae3a6ba..e59c6c7b 100644
--- a/python/pyspark/ml/wrapper.py
+++ b/python/pyspark/ml/wrapper.py
@@ -23,7 +23,8 @@ if sys.version >= '3':
from pyspark import since
from pyspark import SparkContext
from pyspark.sql import DataFrame
-from pyspark.ml import Estimator, Transformer, Model
+from pyspark.ml import Estimator, Predictor, PredictionModel, Transformer, Model
+from pyspark.ml.base import _PredictorParams
from pyspark.ml.param import Params
from pyspark.ml.param.shared import HasFeaturesCol, HasLabelCol, HasPredictionCol
from pyspark.ml.util import _jvm
@@ -377,63 +378,20 @@ class JavaModel(JavaTransformer, Model):
@inherit_doc
-class _JavaPredictorParams(HasLabelCol, HasFeaturesCol, HasPredictionCol):
- """
- Params for :py:class:`JavaPredictor` and :py:class:`JavaPredictorModel`.
-
- .. versionadded:: 3.0.0
- """
- pass
-
-
-@inherit_doc
-class JavaPredictor(JavaEstimator, _JavaPredictorParams):
+class JavaPredictor(Predictor, JavaEstimator, _PredictorParams):
"""
(Private) Java Estimator for prediction tasks (regression and classification).
"""
- @since("3.0.0")
- def setLabelCol(self, value):
- """
- Sets the value of :py:attr:`labelCol`.
- """
- return self._set(labelCol=value)
-
- @since("3.0.0")
- def setFeaturesCol(self, value):
- """
- Sets the value of :py:attr:`featuresCol`.
- """
- return self._set(featuresCol=value)
-
- @since("3.0.0")
- def setPredictionCol(self, value):
- """
- Sets the value of :py:attr:`predictionCol`.
- """
- return self._set(predictionCol=value)
+ __metaclass__ = ABCMeta
@inherit_doc
-class JavaPredictionModel(JavaModel, _JavaPredictorParams):
+class JavaPredictionModel(PredictionModel, JavaModel, _PredictorParams):
"""
(Private) Java Model for prediction tasks (regression and classification).
"""
- @since("3.0.0")
- def setFeaturesCol(self, value):
- """
- Sets the value of :py:attr:`featuresCol`.
- """
- return self._set(featuresCol=value)
-
- @since("3.0.0")
- def setPredictionCol(self, value):
- """
- Sets the value of :py:attr:`predictionCol`.
- """
- return self._set(predictionCol=value)
-
@property
@since("2.1.0")
def numFeatures(self):
---------------------------------------------------------------------
To unsubscribe, e-mail: commits-unsubscribe@spark.apache.org
For additional commands, e-mail: commits-help@spark.apache.org