You are viewing a plain text version of this content. The canonical link for it is here.
Posted to commits@mxnet.apache.org by GitBox <gi...@apache.org> on 2018/03/30 23:10:17 UTC
[GitHub] cjolivier01 closed pull request #10339: [WIP] Refactor engine, remove execution graphs
cjolivier01 closed pull request #10339: [WIP] Refactor engine, remove execution graphs
URL: https://github.com/apache/incubator-mxnet/pull/10339
This is a PR merged from a forked repository.
As GitHub hides the original diff on merge, it is displayed below for
the sake of provenance:
As this is a foreign pull request (from a fork), the diff is supplied
below (as it won't show otherwise due to GitHub magic):
diff --git a/CMakeLists.txt b/CMakeLists.txt
index 116de37fb85..06517c20342 100644
--- a/CMakeLists.txt
+++ b/CMakeLists.txt
@@ -15,7 +15,7 @@ mxnet_option(USE_NCCL "Use NVidia NCCL with CUDA" OFF)
mxnet_option(USE_OPENCV "Build with OpenCV support" ON)
mxnet_option(USE_OPENMP "Build with Openmp support" ON)
mxnet_option(USE_CUDNN "Build with cudnn support" ON) # one could set CUDNN_ROOT for search path
-mxnet_option(USE_SSE "Build with x86 SSE instruction support" ON)
+mxnet_option(USE_SSE "Build with x86 SSE instruction support" AUTO)
mxnet_option(USE_LAPACK "Build with lapack support" ON IF NOT MSVC)
mxnet_option(USE_MKL_IF_AVAILABLE "Use MKL if found" ON)
mxnet_option(USE_MKLML_MKL "Use MKLDNN variant of MKL (if MKL found)" ON IF USE_MKL_IF_AVAILABLE AND UNIX AND (NOT APPLE))
@@ -97,7 +97,7 @@ else(MSVC)
check_cxx_compiler_flag("-std=c++0x" SUPPORT_CXX0X)
# For cross compilation, we can't rely on the compiler which accepts the flag, but mshadow will
# add platform specific includes not available in other arches
- if(USE_SSE)
+ if(USE_SSE STREQUAL "AUTO")
check_cxx_compiler_flag("-msse2" SUPPORT_MSSE2)
else()
set(SUPPORT_MSSE2 FALSE)
@@ -573,14 +573,7 @@ if(NOT MSVC)
set(CMAKE_C_FLAGS "${CMAKE_C_FLAGS}")
set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -std=c++0x")
else()
- set(CMAKE_CXX_FLAGS_DEBUG "${CMAKE_CXX_FLAGS_DEBUG} /EHsc")
- set(CMAKE_CXX_FLAGS_RELEASE "${CMAKE_CXX_FLAGS_RELEASE} /EHsc /Gy")
- set(CMAKE_CXX_FLAGS_MINSIZEREL "${CMAKE_CXX_FLAGS_MINSIZEREL} /EHsc /Gy")
- set(CMAKE_CXX_FLAGS_RELWITHDEBINFO "${CMAKE_CXX_FLAGS_RELWITHDEBINFO} /EHsc /Gy")
- set(CMAKE_SHARED_LINKER_FLAGS_RELEASE "${CMAKE_SHARED_LINKER_FLAGS_RELEASE} /OPT:REF /OPT:ICF")
- set(CMAKE_SHARED_LINKER_FLAGS_MINSIZEREL "${CMAKE_SHARED_LINKER_FLAGS_MINSIZEREL} /OPT:REF /OPT:ICF")
- set(CMAKE_SHARED_LINKER_FLAGS_RELWITHDEBINFO "${CMAKE_SHARED_LINKER_FLAGS_RELWITHDEBINFO} /OPT:REF /OPT:ICF")
-
+ set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} /EHsc")
endif()
set(MXNET_INSTALL_TARGETS mxnet)
@@ -597,8 +590,8 @@ endif()
if(USE_CUDA)
if(FIRST_CUDA AND MSVC)
- target_compile_options(mxnet PUBLIC "$<$<CONFIG:DEBUG>:-Xcompiler=-MTd -Gy>")
- target_compile_options(mxnet PUBLIC "$<$<CONFIG:RELEASE>:-Xcompiler=-MT -Gy>")
+ target_compile_options(mxnet PUBLIC "$<$<CONFIG:DEBUG>:-Xcompiler=-MTd>")
+ target_compile_options(mxnet PUBLIC "$<$<CONFIG:RELEASE>:-Xcompiler=-MT>")
endif()
endif()
if(USE_DIST_KVSTORE)
diff --git a/Jenkinsfile b/Jenkinsfile
index b7be68c6f73..d38c8b5cb9a 100644
--- a/Jenkinsfile
+++ b/Jenkinsfile
@@ -25,7 +25,7 @@ mx_lib = 'lib/libmxnet.so, lib/libmxnet.a, 3rdparty/dmlc-core/libdmlc.a, 3rdpart
// mxnet cmake libraries, in cmake builds we do not produce a libnvvm static library by default.
mx_cmake_lib = 'build/libmxnet.so, build/libmxnet.a, build/3rdparty/dmlc-core/libdmlc.a, build/tests/mxnet_unit_tests, build/3rdparty/openmp/runtime/src/libomp.so'
mx_cmake_mkldnn_lib = 'build/libmxnet.so, build/libmxnet.a, build/3rdparty/dmlc-core/libdmlc.a, build/tests/mxnet_unit_tests, build/3rdparty/openmp/runtime/src/libomp.so, build/3rdparty/mkldnn/src/libmkldnn.so, build/3rdparty/mkldnn/src/libmkldnn.so.0'
-mx_mkldnn_lib = 'lib/libmxnet.so, lib/libmxnet.a, lib/libiomp5.so, lib/libmkldnn.so.0, lib/libmklml_intel.so, 3rdparty/dmlc-core/libdmlc.a, 3rdparty/nnvm/lib/libnnvm.a'
+mx_mkldnn_lib = 'lib/libmxnet.so, lib/libmxnet.a, lib/libiomp5.so, lib/libmklml_gnu.so, lib/libmkldnn.so, lib/libmkldnn.so.0, lib/libmklml_intel.so, 3rdparty/dmlc-core/libdmlc.a, 3rdparty/nnvm/lib/libnnvm.a'
// command to start a docker container
docker_run = 'tests/ci_build/ci_build.sh'
// timeout in minutes
diff --git a/Makefile b/Makefile
index dba649f7311..930499c66f4 100644
--- a/Makefile
+++ b/Makefile
@@ -432,9 +432,9 @@ lib/libmxnet.so: $(ALLX_DEP)
-Wl,${WHOLE_ARCH} $(filter %libnnvm.a, $^) -Wl,${NO_WHOLE_ARCH}
ifeq ($(USE_MKLDNN), 1)
ifeq ($(UNAME_S), Darwin)
- install_name_tool -change '@rpath/libmklml.dylib' '@loader_path/libmklml.dylib' $@
- install_name_tool -change '@rpath/libiomp5.dylib' '@loader_path/libiomp5.dylib' $@
- install_name_tool -change '@rpath/libmkldnn.0.dylib' '@loader_path/libmkldnn.0.dylib' $@
+ install_name_tool -change '@rpath/libmklml.dylib' '@loader_path/libmklml.dylib' lib/libmxnet.so
+ install_name_tool -change '@rpath/libiomp5.dylib' '@loader_path/libiomp5.dylib' lib/libmxnet.so
+ install_name_tool -change '@rpath/libmkldnn.0.dylib' '@loader_path/libmkldnn.0.dylib' lib/libmxnet.so
endif
endif
diff --git a/benchmark/python/sparse/updater.py b/benchmark/python/sparse/updater.py
deleted file mode 100644
index 72f2bfd04a2..00000000000
--- a/benchmark/python/sparse/updater.py
+++ /dev/null
@@ -1,78 +0,0 @@
-# Licensed to the Apache Software Foundation (ASF) under one
-# or more contributor license agreements. See the NOTICE file
-# distributed with this work for additional information
-# regarding copyright ownership. The ASF licenses this file
-# to you under the Apache License, Version 2.0 (the
-# "License"); you may not use this file except in compliance
-# with the License. You may obtain a copy of the License at
-#
-# http://www.apache.org/licenses/LICENSE-2.0
-#
-# Unless required by applicable law or agreed to in writing,
-# software distributed under the License is distributed on an
-# "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY
-# KIND, either express or implied. See the License for the
-# specific language governing permissions and limitations
-# under the License.
-
-import time
-import mxnet as mx
-from mxnet.ndarray.sparse import adam_update
-import numpy as np
-import argparse
-
-mx.random.seed(0)
-np.random.seed(0)
-
-parser = argparse.ArgumentParser(description='Benchmark adam updater')
-parser.add_argument('--dim-in', type=int, default=240000, help='weight.shape[0]')
-parser.add_argument('--dim-out', type=int, default=512, help='weight.shape[1]')
-parser.add_argument('--nnr', type=int, default=5000, help='grad.indices.shape[0]')
-parser.add_argument('--repeat', type=int, default=1000, help='num repeat')
-parser.add_argument('--dense-grad', action='store_true',
- help='if set to true, both gradient and weight are dense.')
-parser.add_argument('--dense-state', action='store_true',
- help='if set to true, states are dense, indicating standard update')
-parser.add_argument('--cpu', action='store_true')
-
-
-args = parser.parse_args()
-dim_in = args.dim_in
-dim_out = args.dim_out
-nnr = args.nnr
-ctx = mx.cpu() if args.cpu else mx.gpu()
-
-ones = mx.nd.ones((dim_in, dim_out), ctx=ctx)
-
-if not args.dense_grad:
- weight = ones.tostype('row_sparse')
- indices = np.arange(dim_in)
- np.random.shuffle(indices)
- indices = np.unique(indices[:nnr])
- indices = mx.nd.array(indices, ctx=ctx)
- grad = mx.nd.sparse.retain(weight, indices)
-else:
- weight = ones.copy()
- grad = ones.copy()
-
-if args.dense_state:
- mean = ones.copy()
-else:
- mean = ones.tostype('row_sparse')
-
-var = mean.copy()
-
-# warmup
-for i in range(10):
- adam_update(weight, grad, mean, var, out=weight, lr=1, wd=0, beta1=0.9,
- beta2=0.99, rescale_grad=0.5, epsilon=1e-8)
-weight.wait_to_read()
-
-# measure speed
-a = time.time()
-for i in range(args.repeat):
- adam_update(weight, grad, mean, var, out=weight, lr=1, wd=0, beta1=0.9,
- beta2=0.99, rescale_grad=0.5, epsilon=1e-8)
-weight.wait_to_read()
-b = time.time()
-print(b - a)
diff --git a/docs/faq/index.md b/docs/faq/index.md
index 098d37f5fc0..099cd509b14 100644
--- a/docs/faq/index.md
+++ b/docs/faq/index.md
@@ -56,8 +56,6 @@ and full working examples, visit the [tutorials section](../tutorials/index.md).
* [How do I create new operators in MXNet?](http://mxnet.io/faq/new_op.html)
-* [How do I implement sparse operators in MXNet backend?](https://cwiki.apache.org/confluence/display/MXNET/A+Guide+to+Implementing+Sparse+Operators+in+MXNet+Backend)
-
* [How do I contribute an example or tutorial?](https://github.com/apache/incubator-mxnet/tree/master/example#contributing)
* [How do I set MXNet's environmental variables?](http://mxnet.io/faq/env_var.html)
diff --git a/docs/install/osx_setup.md b/docs/install/osx_setup.md
index 4d979b3dccf..c1fa0fcd7f1 100644
--- a/docs/install/osx_setup.md
+++ b/docs/install/osx_setup.md
@@ -1,4 +1,4 @@
-# Installing MXNet from source on OS X (Mac)
+# Installing MXNet froum source on OS X (Mac)
**NOTE:** For prebuild MXNet with Python installation, please refer to the [new install guide](http://mxnet.io/install/index.html).
@@ -65,8 +65,8 @@ Install the dependencies, required for MXNet, with the following commands:
brew install openblas
brew tap homebrew/core
brew install opencv
- # Get pip
- easy_install pip
+ # For getting pip
+ brew install python
# For visualization of network graphs
pip install graphviz
# Jupyter notebook
@@ -167,12 +167,6 @@ You might want to add this command to your ```~/.bashrc``` file. If you do, you
For more details about installing and using MXNet with Julia, see the [MXNet Julia documentation](http://dmlc.ml/MXNet.jl/latest/user-guide/install/).
## Install the MXNet Package for Scala
-
-If you haven't installed maven yet, you need to install it now (required by the makefile):
-```bash
- brew install maven
-```
-
Before you build MXNet for Scala from source code, you must complete [building the shared library](#build-the-shared-library). After you build the shared library, run the following command from the MXNet source root directory to build the MXNet Scala package:
```bash
diff --git a/docs/mxdoc.py b/docs/mxdoc.py
index 7f567f0b8d0..907ec7cc57f 100644
--- a/docs/mxdoc.py
+++ b/docs/mxdoc.py
@@ -265,11 +265,9 @@ def _get_python_block_output(src, global_dict, local_dict):
ret_status = False
return (ret_status, s.getvalue()+err)
-def _get_jupyter_notebook(lang, all_lines):
+def _get_jupyter_notebook(lang, lines):
cells = []
- # Exclude lines containing <!--notebook-skip-line-->
- filtered_lines = [line for line in all_lines if "<!--notebook-skip-line-->" not in line]
- for in_code, blk_lang, lines in _get_blocks(filtered_lines):
+ for in_code, blk_lang, lines in _get_blocks(lines):
if blk_lang != lang:
in_code = False
src = '\n'.join(lines)
diff --git a/docs/tutorials/index.md b/docs/tutorials/index.md
index 8a597e95bfb..3eff299d778 100644
--- a/docs/tutorials/index.md
+++ b/docs/tutorials/index.md
@@ -119,8 +119,6 @@ The Gluon and Module tutorials are in Python, but you can also find a variety of
- [Simple autograd example](http://mxnet.incubator.apache.org/tutorials/gluon/autograd.html)
-- [Inference using an ONNX model](http://mxnet.incubator.apache.org/tutorials/onnx/inference_on_onnx_model.html)
-
</div> <!--end of applications-->
</div> <!--end of gluon-->
diff --git a/docs/tutorials/onnx/inference_on_onnx_model.md b/docs/tutorials/onnx/inference_on_onnx_model.md
deleted file mode 100644
index 182a2ae74cd..00000000000
--- a/docs/tutorials/onnx/inference_on_onnx_model.md
+++ /dev/null
@@ -1,273 +0,0 @@
-
-# Running inference on MXNet/Gluon from an ONNX model
-
-[Open Neural Network Exchange (ONNX)](https://github.com/onnx/onnx) provides an open source format for AI models. It defines an extensible computation graph model, as well as definitions of built-in operators and standard data types.
-
-In this tutorial we will:
-
-- learn how to load a pre-trained .onnx model file into MXNet/Gluon
-- learn how to test this model using the sample input/output
-- learn how to test the model on custom images
-
-## Pre-requisite
-
-To run the tutorial you will need to have installed the following python modules:
-- [MXNet](http://mxnet.incubator.apache.org/install/index.html)
-- [onnx](https://github.com/onnx/onnx) (follow the install guide)
-- [onnx-mxnet](https://github.com/onnx/onnx-mxnet)
-- matplotlib
-- wget
-
-
-```python
-import numpy as np
-import mxnet as mx
-from mxnet.contrib import onnx as onnx_mxnet
-from mxnet import gluon, nd
-%matplotlib inline
-import matplotlib.pyplot as plt
-import tarfile, os
-import wget
-import json
-```
-
-### Downloading supporting files
-These are images and a vizualisation script
-
-
-```python
-image_folder = "images"
-utils_file = "utils.py" # contain utils function to plot nice visualization
-image_net_labels_file = "image_net_labels.json"
-images = ['apron', 'hammerheadshark', 'dog', 'wrench', 'dolphin', 'lotus']
-base_url = "https://raw.githubusercontent.com/dmlc/web-data/master/mxnet/doc/tutorials/onnx/{}?raw=true"
-
-if not os.path.isdir(image_folder):
- os.makedirs(image_folder)
- for image in images:
- wget.download(base_url.format("{}/{}.jpg".format(image_folder, image)), image_folder)
-if not os.path.isfile(utils_file):
- wget.download(base_url.format(utils_file))
-if not os.path.isfile(image_net_labels_file):
- wget.download(base_url.format(image_net_labels_file))
-```
-
-
-```python
-from utils import *
-```
-
-## Downloading a model from the ONNX model zoo
-
-We download a pre-trained model, in our case the [vgg16](https://arxiv.org/abs/1409.1556) model, trained on [ImageNet](http://www.image-net.org/) from the [ONNX model zoo](https://github.com/onnx/models). The model comes packaged in an archive `tar.gz` file containing an `model.onnx` model file and some sample input/output data.
-
-
-```python
-base_url = "https://s3.amazonaws.com/download.onnx/models/"
-current_model = "vgg16"
-model_folder = "model"
-archive = "{}.tar.gz".format(current_model)
-archive_file = os.path.join(model_folder, archive)
-url = "{}{}".format(base_url, archive)
-```
-
-Create the model folder and download the zipped model
-
-
-```python
-if not os.path.isdir(model_folder):
- os.makedirs(model_folder)
-if not os.path.isfile(archive_file):
- wget.download(url, model_folder)
-```
-
-Extract the model
-
-
-```python
-if not os.path.isdir(os.path.join(model_folder, current_model)):
- tar = tarfile.open(archive_file, "r:gz")
- tar.extractall(model_folder)
- tar.close()
-```
-
-The models have been pre-trained on ImageNet, let's load the label mapping of the 1000 classes.
-
-
-```python
-categories = json.load(open(image_net_labels_file, 'r'))
-```
-
-## Loading the model into MXNet Gluon
-
-
-```python
-onnx_path = os.path.join(model_folder, current_model, "model.onnx")
-```
-
-We get the symbol and parameter objects
-
-
-```python
-sym, arg_params, aux_params = onnx_mxnet.import_model(onnx_path)
-```
-
-We pick a context, CPU or GPU
-
-
-```python
-ctx = mx.cpu()
-```
-
-And load them into a MXNet Gluon symbol block. For ONNX models the default input name is `input_0`.
-
-
-```python
-net = gluon.nn.SymbolBlock(outputs=sym, inputs=mx.sym.var('input_0'))
-net_params = net.collect_params()
-for param in arg_params:
- if param in net_params:
- net_params[param]._load_init(arg_params[param], ctx=ctx)
-for param in aux_params:
- if param in net_params:
- net_params[param]._load_init(aux_params[param], ctx=ctx)
-```
-
-We can now cache the computational graph through [hybridization](https://mxnet.incubator.apache.org/tutorials/gluon/hybrid.html) to gain some performance
-
-
-
-```python
-net.hybridize()
-```
-
-## Test using sample inputs and outputs
-The model comes with sample input/output we can use to test that whether model is correctly loaded
-
-
-```python
-numpy_path = os.path.join(model_folder, current_model, 'test_data_0.npz')
-sample = np.load(numpy_path, encoding='bytes')
-inputs = sample['inputs']
-outputs = sample['outputs']
-```
-
-
-```python
-print("Input format: {}".format(inputs[0].shape))
-print("Output format: {}".format(outputs[0].shape))
-```
-
-`Input format: (1, 3, 224, 224)` <!--notebook-skip-line-->
-
-
-`Output format: (1, 1000)` <!--notebook-skip-line-->
-
-
-
-We can visualize the network (requires graphviz installed)
-
-
-```python
-mx.visualization.plot_network(sym, node_attrs={"shape":"oval","fixedsize":"false"})
-```
-
-
-
-
-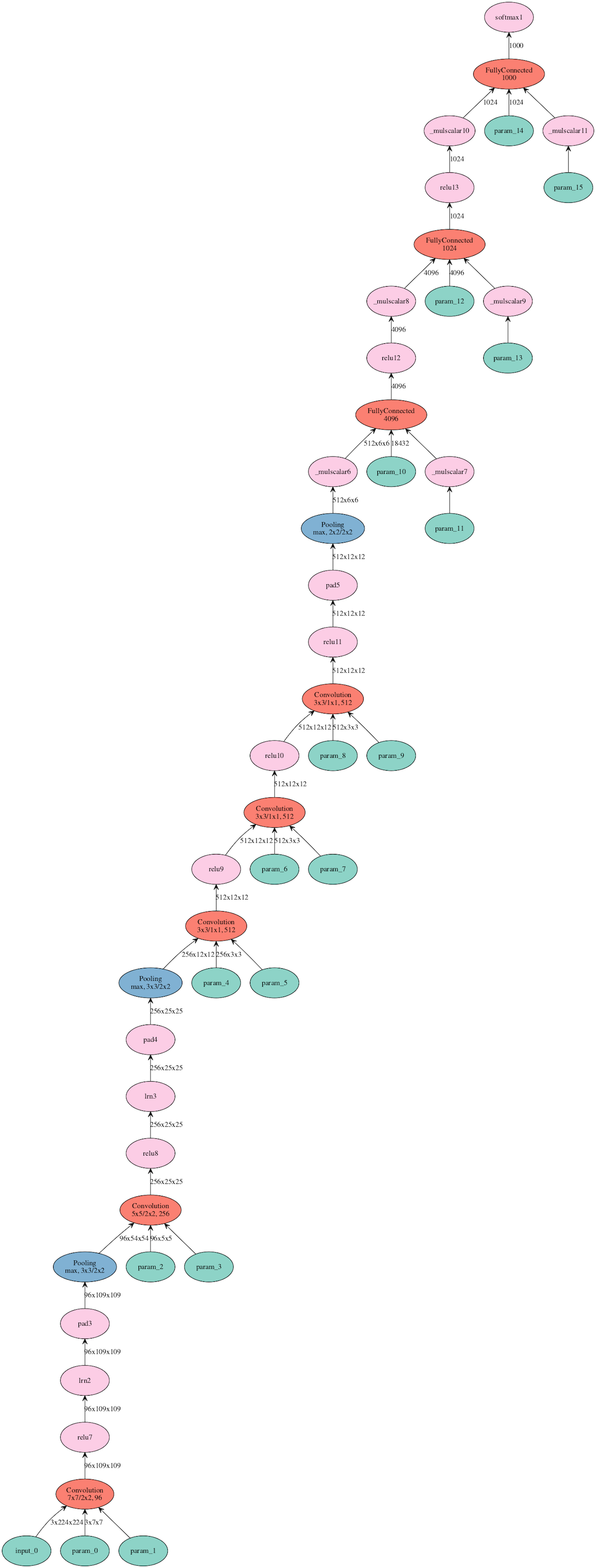<!--notebook-skip-line-->
-
-
-
-This is a helper function to run M batches of data of batch-size N through the net and collate the outputs into an array of shape (K, 1000) where K=MxN is the total number of examples (mumber of batches x batch-size) run through the network.
-
-
-```python
-def run_batch(net, data):
- results = []
- for batch in data:
- outputs = net(batch)
- results.extend([o for o in outputs.asnumpy()])
- return np.array(results)
-```
-
-
-```python
-result = run_batch(net, nd.array([inputs[0]], ctx))
-```
-
-
-```python
-print("Loaded model and sample output predict the same class: {}".format(np.argmax(result) == np.argmax(outputs[0])))
-```
-
-Loaded model and sample output predict the same class: True <!--notebook-skip-line-->
-
-
-Good the sample output and our prediction match, now we can run against real data
-
-## Test using real images
-
-
-```python
-TOP_P = 3 # How many top guesses we show in the visualization
-```
-
-
-Transform function to set the data into the format the network expects, (N, 3, 224, 224) where N is the batch size.
-
-
-```python
-def transform(img):
- return np.expand_dims(np.transpose(img, (2,0,1)),axis=0).astype(np.float32)
-```
-
-
-We load two sets of images in memory
-
-
-```python
-image_net_images = [plt.imread('images/{}.jpg'.format(path)) for path in ['apron', 'hammerheadshark','dog']]
-caltech101_images = [plt.imread('images/{}.jpg'.format(path)) for path in ['wrench', 'dolphin','lotus']]
-images = image_net_images + caltech101_images
-```
-
-And run them as a batch through the network to get the predictions
-
-```python
-batch = nd.array(np.concatenate([transform(img) for img in images], axis=0), ctx=ctx)
-result = run_batch(net, [batch])
-```
-
-
-```python
-plot_predictions(image_net_images, result[:3], categories, TOP_P)
-```
-
-
-<!--notebook-skip-line-->
-
-
-**Well done!** Looks like it is doing a pretty good job at classifying pictures when the category is a ImageNet label
-
-Let's now see the results on the 3 other images
-
-
-```python
-plot_predictions(caltech101_images, result[3:7], categories, TOP_P)
-```
-
-
-<!--notebook-skip-line-->
-
-
-**Hmm, not so good...** Even though predictions are close, they are not accurate, which is due to the fact that the ImageNet dataset does not contain `wrench`, `dolphin`, or `lotus` categories and our network has been trained on ImageNet.
-
-Lucky for us, the [Caltech101 dataset](http://www.vision.caltech.edu/Image_Datasets/Caltech101/) has them, let's see how we can fine-tune our network to classify these categories correctly.
-
-We show that in our next tutorials:
-
-- Fine-tuning a ONNX Model using the modern imperative MXNet/Gluon API(Coming soon)
-- Fine-tuning a ONNX Model using the symbolic MXNet/Module API(Coming soon)
-
-<!-- INSERT SOURCE DOWNLOAD BUTTONS -->
diff --git a/example/README.md b/example/README.md
index 1ad66e942de..49484a09e97 100644
--- a/example/README.md
+++ b/example/README.md
@@ -38,12 +38,6 @@ The site expects the format to be markdown, so export your notebook as a .md via
<!-- INSERT SOURCE DOWNLOAD BUTTONS -->
```
-If you want some lines to show-up in the markdown but not in the generated notebooks, add this comment `<!--notebook-skip-line-->` after your ``. Like this:
-```
-<!--notebook-skip-line-->
-```
-Typically when you have a `plt.imshow()` you want the image tag `[png](img.png)` in the `.md` but not in the downloaded notebook as the user will re-generate the plot at run-time.
-
## <a name="list-of-examples"></a>List of examples
### <a name="language-binding-examples"></a>Languages Binding Examples
diff --git a/example/gluon/image_classification.py b/example/gluon/image_classification.py
index a67a31790a0..9acfda51d17 100644
--- a/example/gluon/image_classification.py
+++ b/example/gluon/image_classification.py
@@ -22,7 +22,6 @@
import mxnet as mx
from mxnet import gluon
-from mxnet import profiler
from mxnet.gluon import nn
from mxnet.gluon.model_zoo import vision as models
from mxnet import autograd as ag
@@ -97,7 +96,6 @@
parser.add_argument('--profile', action='store_true',
help='Option to turn on memory profiling for front-end, '\
'and prints out the memory usage by python function at the end.')
-parser.add_argument('--builtin-profiler', type=int, default=0, help='Enable built-in profiler (0=off, 1=on)')
opt = parser.parse_args()
# global variables
@@ -196,9 +194,6 @@ def train(opt, ctx):
kvstore = kv)
loss = gluon.loss.SoftmaxCrossEntropyLoss()
-
- total_time = 0
- num_epochs = 0
best_acc = [0]
for epoch in range(opt.start_epoch, opt.epochs):
trainer = update_learning_rate(opt.lr, trainer, epoch, opt.lr_factor, lr_steps)
@@ -228,29 +223,16 @@ def train(opt, ctx):
epoch, i, batch_size/(time.time()-btic), name[0], acc[0], name[1], acc[1]))
btic = time.time()
- epoch_time = time.time()-tic
-
- # First epoch will usually be much slower than the subsequent epics,
- # so don't factor into the average
- if num_epochs > 0:
- total_time = total_time + epoch_time
- num_epochs = num_epochs + 1
-
name, acc = metric.get()
logger.info('[Epoch %d] training: %s=%f, %s=%f'%(epoch, name[0], acc[0], name[1], acc[1]))
- logger.info('[Epoch %d] time cost: %f'%(epoch, epoch_time))
+ logger.info('[Epoch %d] time cost: %f'%(epoch, time.time()-tic))
name, val_acc = test(ctx, val_data)
logger.info('[Epoch %d] validation: %s=%f, %s=%f'%(epoch, name[0], val_acc[0], name[1], val_acc[1]))
# save model if meet requirements
save_checkpoint(epoch, val_acc[0], best_acc)
- if num_epochs > 1:
- print('Average epoch time: {}'.format(float(total_time)/(num_epochs - 1)))
def main():
- if opt.builtin_profiler > 0:
- profiler.set_config(profile_all=True, aggregate_stats=True)
- profiler.set_state('run')
if opt.mode == 'symbolic':
data = mx.sym.var('data')
out = net(data)
@@ -272,9 +254,6 @@ def main():
if opt.mode == 'hybrid':
net.hybridize()
train(opt, context)
- if opt.builtin_profiler > 0:
- profiler.set_state('stop')
- print(profiler.dumps())
if __name__ == '__main__':
if opt.profile:
diff --git a/example/onnx/super_resolution.py b/example/onnx/super_resolution.py
index f7c7886d0df..1392b77715c 100644
--- a/example/onnx/super_resolution.py
+++ b/example/onnx/super_resolution.py
@@ -37,9 +37,9 @@ def import_onnx():
download(model_url, 'super_resolution.onnx')
LOGGER.info("Converting onnx format to mxnet's symbol and params...")
- sym, arg_params, aux_params = onnx_mxnet.import_model('super_resolution.onnx')
+ sym, params = onnx_mxnet.import_model('super_resolution.onnx')
LOGGER.info("Successfully Converted onnx format to mxnet's symbol and params...")
- return sym, arg_params, aux_params
+ return sym, params
def get_test_image():
"""Download and process the test image"""
@@ -53,12 +53,12 @@ def get_test_image():
input_image = np.array(img_y)[np.newaxis, np.newaxis, :, :]
return input_image, img_cb, img_cr
-def perform_inference(sym, arg_params, aux_params, input_img, img_cb, img_cr):
+def perform_inference(sym, params, input_img, img_cb, img_cr):
"""Perform inference on image using mxnet"""
# create module
mod = mx.mod.Module(symbol=sym, data_names=['input_0'], label_names=None)
mod.bind(for_training=False, data_shapes=[('input_0', input_img.shape)])
- mod.set_params(arg_params=arg_params, aux_params=aux_params)
+ mod.set_params(arg_params=params, aux_params=None)
# run inference
batch = namedtuple('Batch', ['data'])
@@ -79,6 +79,6 @@ def perform_inference(sym, arg_params, aux_params, input_img, img_cb, img_cr):
return result_img
if __name__ == '__main__':
- MX_SYM, MX_ARG_PARAM, MX_AUX_PARAM = import_onnx()
+ MX_SYM, MX_PARAM = import_onnx()
INPUT_IMG, IMG_CB, IMG_CR = get_test_image()
- perform_inference(MX_SYM, MX_ARG_PARAM, MX_AUX_PARAM, INPUT_IMG, IMG_CB, IMG_CR)
+ perform_inference(MX_SYM, MX_PARAM, INPUT_IMG, IMG_CB, IMG_CR)
diff --git a/include/mxnet/ndarray.h b/include/mxnet/ndarray.h
index b8b7f20fcf3..e6e7468a1f4 100644
--- a/include/mxnet/ndarray.h
+++ b/include/mxnet/ndarray.h
@@ -279,12 +279,12 @@ class NDArray {
CHECK_EQ(aux_shape(rowsparse::kIdx)[0], storage_shape()[0])
<< "inconsistent storage shape " << storage_shape()
<< " vs. aux shape " << aux_shape(rowsparse::kIdx);
- return aux_shape(rowsparse::kIdx).Size() != 0;
+ return aux_shape(0).Size() != 0;
} else if (stype == kCSRStorage) {
CHECK_EQ(aux_shape(csr::kIdx)[0], storage_shape()[0])
<< "inconsistent storage shape " << storage_shape()
<< " vs. aux shape " << aux_shape(csr::kIdx);
- return aux_shape(csr::kIdx).Size() != 0;
+ return aux_shape(0).Size() != 0;
} else {
LOG(FATAL) << "Unknown storage type";
}
@@ -623,6 +623,18 @@ class NDArray {
mkldnn::memory *CreateMKLDNNData(
const mkldnn::memory::primitive_desc &desc);
+ /*
+ * Reorder the memory to the specified layout.
+ */
+ void MKLDNNDataReorder(const mkldnn::memory::primitive_desc &desc) {
+ CHECK_EQ(storage_type(), kDefaultStorage);
+ ptr_->MKLDNNDataReorder(desc);
+ }
+ void Reorder2Default() {
+ CHECK_EQ(storage_type(), kDefaultStorage);
+ ptr_->Reorder2Default();
+ }
+
/*
* These are the async version of the methods above.
* It changes the layout of this NDArray, but it happens after all accesses to
diff --git a/prepare_mkldnn.sh b/prepare_mkldnn.sh
index df5e9b99656..50552eb22f6 100755
--- a/prepare_mkldnn.sh
+++ b/prepare_mkldnn.sh
@@ -73,13 +73,9 @@ if [ ! -z "$HOME_MKLDNN" ]; then
fi
if [ $OSTYPE == "darwin16" ]; then
- OMP_LIBFILE="$MKLDNN_INSTALLDIR/lib/libiomp5.dylib"
- MKLML_LIBFILE="$MKLDNN_INSTALLDIR/lib/libmklml.dylib"
- MKLDNN_LIBFILE="$MKLDNN_INSTALLDIR/lib/libmkldnn.0.dylib"
+ MKLDNN_LIBFILE="$MKLDNN_INSTALLDIR/lib/libmkldnn.dylib"
else
- OMP_LIBFILE="$MKLDNN_INSTALLDIR/lib/libiomp5.so"
- MKLML_LIBFILE="$MKLDNN_INSTALLDIR/lib/libmklml_intel.so"
- MKLDNN_LIBFILE="$MKLDNN_INSTALLDIR/lib/libmkldnn.so.0"
+ MKLDNN_LIBFILE="$MKLDNN_INSTALLDIR/lib/libmkldnn.so"
fi
if [ -z $MKLDNNROOT ]; then
@@ -107,9 +103,7 @@ if [ ! -f $MKLDNN_LIBFILE ]; then
make -C $MKLDNN_BUILDDIR install >&2
rm -rf $MKLDNN_BUILDDIR
mkdir -p $MKLDNN_LIBDIR
- cp $OMP_LIBFILE $MKLDNN_LIBDIR
- cp $MKLML_LIBFILE $MKLDNN_LIBDIR
- cp $MKLDNN_LIBFILE $MKLDNN_LIBDIR
+ cp $MKLDNN_INSTALLDIR/lib/* $MKLDNN_LIBDIR
fi
MKLDNNROOT=$MKLDNN_INSTALLDIR
fi
diff --git a/python/mxnet/contrib/onnx/_import/import_helper.py b/python/mxnet/contrib/onnx/_import/import_helper.py
index 175c2fb6a00..80541ec3577 100644
--- a/python/mxnet/contrib/onnx/_import/import_helper.py
+++ b/python/mxnet/contrib/onnx/_import/import_helper.py
@@ -27,7 +27,7 @@
from .op_translations import global_avgpooling, global_maxpooling, linalg_gemm
from .op_translations import sigmoid, pad, relu, matrix_multiplication, batch_norm
from .op_translations import dropout, local_response_norm, conv, deconv
-from .op_translations import reshape, cast, split, _slice, transpose, squeeze, flatten
+from .op_translations import reshape, cast, split, _slice, transpose, squeeze
from .op_translations import reciprocal, squareroot, power, exponent, _log
from .op_translations import reduce_max, reduce_mean, reduce_min, reduce_sum
from .op_translations import reduce_prod, avg_pooling, max_pooling
@@ -83,7 +83,6 @@
'Slice' : _slice,
'Transpose' : transpose,
'Squeeze' : squeeze,
- 'Flatten' : flatten,
#Powers
'Reciprocal' : reciprocal,
'Sqrt' : squareroot,
diff --git a/python/mxnet/contrib/onnx/_import/import_model.py b/python/mxnet/contrib/onnx/_import/import_model.py
index d8d32a96a21..1df429b4690 100644
--- a/python/mxnet/contrib/onnx/_import/import_model.py
+++ b/python/mxnet/contrib/onnx/_import/import_model.py
@@ -46,5 +46,5 @@ def import_model(model_file):
except ImportError:
raise ImportError("Onnx and protobuf need to be installed")
model_proto = onnx.load(model_file)
- sym, arg_params, aux_params = graph.from_onnx(model_proto.graph)
- return sym, arg_params, aux_params
+ sym, params = graph.from_onnx(model_proto.graph)
+ return sym, params
diff --git a/python/mxnet/contrib/onnx/_import/import_onnx.py b/python/mxnet/contrib/onnx/_import/import_onnx.py
index 037790c8080..56181c777be 100644
--- a/python/mxnet/contrib/onnx/_import/import_onnx.py
+++ b/python/mxnet/contrib/onnx/_import/import_onnx.py
@@ -61,12 +61,12 @@ def _convert_operator(self, node_name, op_name, attrs, inputs):
raise NotImplementedError("Operator {} not implemented.".format(op_name))
if isinstance(op_name, string_types):
new_op = getattr(symbol, op_name, None)
- if not new_op:
- raise RuntimeError("Unable to map op_name {} to sym".format(op_name))
if node_name is None:
mxnet_sym = new_op(*inputs, **new_attrs)
else:
mxnet_sym = new_op(name=node_name, *inputs, **new_attrs)
+ if not mxnet_sym:
+ raise RuntimeError("Unable to map op_name {} to sym".format(op_name))
return mxnet_sym
return op_name
@@ -110,10 +110,6 @@ def from_onnx(self, graph):
self._nodes[name_input] = symbol.Variable(name=name_input)
self._renames[i.name] = name_input
- # For storing arg and aux params for the graph.
- auxDict = {}
- argDict = {}
-
# constructing nodes, nodes are stored as directed acyclic graph
# converting NodeProto message
for node in graph.node:
@@ -124,24 +120,19 @@ def from_onnx(self, graph):
inputs = [self._nodes[self._renames.get(i, i)] for i in node.input]
mxnet_sym = self._convert_operator(node_name, op_name, onnx_attr, inputs)
- for k, i in zip(list(node.output), range(len(mxnet_sym.list_outputs()))):
+ assert len(node.output) == len(mxnet_sym.list_outputs()), (
+ "Output dimension mismatch between the onnx operator and the mxnet symbol " +
+ "{} vs {} for the operator - {}.".format(
+ len(node.output), len(mxnet_sym.list_outputs()), op_name))
+ for k, i in zip(list(node.output), range(len(node.output))):
self._nodes[k] = mxnet_sym[i]
-
- # splitting params into args and aux params
- for args in mxnet_sym.list_arguments():
- if args in self._params:
- argDict.update({args: nd.array(self._params[args])})
- for aux in mxnet_sym.list_auxiliary_states():
- if aux in self._params:
- auxDict.update({aux: nd.array(self._params[aux])})
-
# now return the outputs
out = [self._nodes[i.name] for i in graph.output]
if len(out) > 1:
out = symbol.Group(out)
else:
out = out[0]
- return out, argDict, auxDict
+ return out, self._params
def _parse_array(self, tensor_proto):
"""Grab data in TensorProto and convert to numpy array."""
diff --git a/python/mxnet/contrib/onnx/_import/op_translations.py b/python/mxnet/contrib/onnx/_import/op_translations.py
index de341321785..a67c18199eb 100644
--- a/python/mxnet/contrib/onnx/_import/op_translations.py
+++ b/python/mxnet/contrib/onnx/_import/op_translations.py
@@ -164,14 +164,10 @@ def matrix_multiplication(attrs, inputs, cls):
def batch_norm(attrs, inputs, cls):
"""Batch normalization."""
- new_attrs = translation_utils._fix_attribute_names(attrs, {'epsilon' : 'eps',
- 'is_test':'fix_gamma'})
+ new_attrs = translation_utils._fix_attribute_names(attrs, {'epsilon' : 'eps'})
new_attrs = translation_utils._remove_attributes(new_attrs,
- ['spatial', 'consumed_inputs'])
+ ['spatial', 'is_test', 'consumed_inputs'])
new_attrs = translation_utils._add_extra_attributes(new_attrs, {'cudnn_off': 1})
-
- # in test mode "fix_gamma" should be unset.
- new_attrs['fix_gamma'] = 0 if new_attrs['fix_gamma'] == 1 else 1
return 'BatchNorm', new_attrs, inputs
@@ -249,7 +245,7 @@ def global_maxpooling(attrs, inputs, cls):
new_attrs = translation_utils._add_extra_attributes(attrs, {'global_pool': True,
'kernel': (1, 1),
'pool_type': 'max'})
- return 'Pooling', new_attrs, inputs
+ return 'pooling', new_attrs, inputs
def global_avgpooling(attrs, inputs, cls):
@@ -257,49 +253,28 @@ def global_avgpooling(attrs, inputs, cls):
new_attrs = translation_utils._add_extra_attributes(attrs, {'global_pool': True,
'kernel': (1, 1),
'pool_type': 'avg'})
- return 'Pooling', new_attrs, inputs
+ return 'pooling', new_attrs, inputs
def linalg_gemm(attrs, inputs, cls):
"""Performs general matrix multiplication and accumulation"""
- trans_a = 0
- trans_b = 0
- alpha = 1
- beta = 1
- if 'transA' in attrs:
- trans_a = attrs['transA']
- if 'transB' in attrs:
- trans_b = attrs['transB']
- if 'alpha' in attrs:
- alpha = attrs['alpha']
- if 'beta' in attrs:
- beta = attrs['beta']
- flatten_a = symbol.flatten(inputs[0])
- matmul_op = symbol.linalg_gemm2(A=flatten_a, B=inputs[1],
- transpose_a=trans_a, transpose_b=trans_b,
- alpha=alpha)
- gemm_op = symbol.broadcast_add(matmul_op, beta*inputs[2])
new_attrs = translation_utils._fix_attribute_names(attrs, {'transA': 'transpose_a',
'transB': 'transpose_b'})
new_attrs = translation_utils._remove_attributes(new_attrs, ['broadcast'])
- return gemm_op, new_attrs, inputs
+ return translation_utils._fix_gemm('FullyConnected', inputs, new_attrs, cls)
-def local_response_norm(attrs, inputs, cls):
+def local_response_norm(op_name, attrs, inputs):
"""Local Response Normalization."""
new_attrs = translation_utils._fix_attribute_names(attrs,
{'bias': 'knorm',
'size' : 'nsize'})
return 'LRN', new_attrs, inputs
-def dropout(attrs, inputs, cls):
+def dropout(op_name, attrs, inputs):
"""Dropout Regularization."""
- mode = 'training'
- if attrs['is_test'] == 0:
- mode = 'always'
new_attrs = translation_utils._fix_attribute_names(attrs,
{'ratio': 'p'})
new_attrs = translation_utils._remove_attributes(new_attrs, ['is_test'])
- new_attrs = translation_utils._add_extra_attributes(new_attrs, {'mode': mode})
return 'Dropout', new_attrs, inputs
# Changing shape and type.
@@ -310,7 +285,6 @@ def reshape(attrs, inputs, cls):
def cast(attrs, inputs, cls):
""" Cast input to a given dtype"""
new_attrs = translation_utils._fix_attribute_names(attrs, {'to' : 'dtype'})
- new_attrs['dtype'] = new_attrs['dtype'].lower()
return 'cast', new_attrs, inputs
def split(attrs, inputs, cls):
@@ -354,15 +328,6 @@ def squeeze(attrs, inputs, cls):
mxnet_op = symbol.split(mxnet_op, axis=i-1, num_outputs=1, squeeze_axis=1)
return mxnet_op, new_attrs, inputs
-
-def flatten(attrs, inputs, cls):
- """Flattens the input array into a 2-D array by collapsing the higher dimensions."""
- #Mxnet does not have axis support. By default uses axis=1
- if 'axis' in attrs and attrs['axis'] != 1:
- raise RuntimeError("Flatten operator only supports axis=1")
- new_attrs = translation_utils._remove_attributes(attrs, ['axis'])
- return 'Flatten', new_attrs, inputs
-
#Powers
def reciprocal(attrs, inputs, cls):
"""Returns the reciprocal of the argument, element-wise."""
@@ -422,7 +387,8 @@ def avg_pooling(attrs, inputs, cls):
'pads': 'pad',
})
new_attrs = translation_utils._add_extra_attributes(new_attrs,
- {'pooling_convention': 'valid'
+ {'pool_type': 'avg',
+ 'pooling_convention': 'valid'
})
new_op = translation_utils._fix_pooling('avg', inputs, new_attrs)
@@ -436,9 +402,9 @@ def max_pooling(attrs, inputs, cls):
'strides': 'stride',
'pads': 'pad',
})
-
new_attrs = translation_utils._add_extra_attributes(new_attrs,
- {'pooling_convention': 'valid'
+ {'pool_type': 'avg',
+ 'pooling_convention': 'valid'
})
new_op = translation_utils._fix_pooling('max', inputs, new_attrs)
diff --git a/python/mxnet/contrib/onnx/_import/translation_utils.py b/python/mxnet/contrib/onnx/_import/translation_utils.py
index 1d84bd70d7e..0fdef647b50 100644
--- a/python/mxnet/contrib/onnx/_import/translation_utils.py
+++ b/python/mxnet/contrib/onnx/_import/translation_utils.py
@@ -90,51 +90,10 @@ def _fix_pooling(pool_type, inputs, new_attr):
stride = new_attr.get('stride')
kernel = new_attr.get('kernel')
padding = new_attr.get('pad')
-
- # Adding default stride.
- if stride is None:
- stride = (1,) * len(kernel)
-
- # Add padding attr if not provided.
- if padding is None:
- padding = (0,) * len(kernel) * 2
-
- # Mxnet Pad operator supports only 4D/5D tensors.
- # For 1D case, these are the steps:
- # Step 1. Add extra dummy dimension to make it 4D. Adding to axis = 2
- # Step 2. Apply padding to this changed tensor
- # Step 3. Remove the extra dimension added in step 1.
- if len(kernel) == 1:
- dummy_axis = 2
- # setting 0 padding to the new dim to be added.
- padding = (0, padding[0], 0, padding[1])
- pad_width = (0, 0, 0, 0) + _pad_sequence_fix(padding, kernel_dim=2)
-
- # Step 1.
- curr_sym = symbol.expand_dims(inputs[0], axis=dummy_axis)
-
- # Step 2. Common for all tensor sizes
- new_pad_op = symbol.pad(curr_sym, mode='edge', pad_width=pad_width)
-
- # Step 3: Removing extra dim added.
- new_pad_op = symbol.split(new_pad_op, axis=dummy_axis, num_outputs=1, squeeze_axis=1)
- else:
- # For 2D/3D cases:
- # Apply padding
- pad_width = (0, 0, 0, 0) + _pad_sequence_fix(padding, kernel_dim=len(kernel))
- curr_sym = inputs[0]
-
- if pool_type == 'max':
- # For max pool : mode = 'edge', we should replicate the
- # edge values to pad, so that we only include input data values
- # for calculating 'max'
- new_pad_op = symbol.pad(curr_sym, mode='edge', pad_width=pad_width)
- else:
- # For avg pool, we should add 'zeros' for padding so mode='constant'
- new_pad_op = symbol.pad(curr_sym, mode='constant', pad_width=pad_width)
-
- # Apply pooling without pads.
- new_pooling_op = symbol.Pooling(new_pad_op, pool_type=pool_type, stride=stride, kernel=kernel)
+ pad_width = (0, 0, 0, 0) + _pad_sequence_fix(padding, len(kernel))
+ new_pad_op = symbol.pad(inputs[0], mode='constant', pad_width=pad_width)
+ new_pooling_op = symbol.Pooling(new_pad_op, pool_type=pool_type,
+ stride=stride, kernel=kernel)
return new_pooling_op
def _fix_bias(op_name, attrs, num_inputs):
diff --git a/python/mxnet/gluon/parameter.py b/python/mxnet/gluon/parameter.py
index 5a9277b2d63..7dc72433926 100644
--- a/python/mxnet/gluon/parameter.py
+++ b/python/mxnet/gluon/parameter.py
@@ -574,32 +574,13 @@ def get(self, name, **kwargs):
"""
name = self.prefix + name
param = self._get_impl(name)
- if param is None: # pylint: disable=too-many-nested-blocks
+ if param is None:
param = Parameter(name, **kwargs)
self._params[name] = param
else:
for k, v in kwargs.items():
if hasattr(param, k) and getattr(param, k) is not None:
- existing = getattr(param, k)
- if k == 'shape' and len(v) == len(existing):
- inferred_shape = []
- matched = True
- for dim1, dim2 in zip(v, existing):
- if dim1 != dim2 and dim1 * dim2 != 0:
- matched = False
- break
- elif dim1 == dim2:
- inferred_shape.append(dim1)
- elif dim1 == 0:
- inferred_shape.append(dim2)
- else:
- inferred_shape.append(dim1)
-
- if matched:
- param._shape = tuple(inferred_shape)
- continue
-
- assert v is None or v == existing, \
+ assert v is None or v == getattr(param, k), \
"Cannot retrieve Parameter %s because desired attribute " \
"does not match with stored for attribute %s: " \
"desired %s vs stored %s."%(
diff --git a/python/mxnet/initializer.py b/python/mxnet/initializer.py
index 1297c3da9a7..78afa2dbd29 100755
--- a/python/mxnet/initializer.py
+++ b/python/mxnet/initializer.py
@@ -530,9 +530,9 @@ def _init_weight(self, _, arr):
nout = arr.shape[0]
nin = np.prod(arr.shape[1:])
if self.rand_type == "uniform":
- tmp = random.uniform(-1.0, 1.0, shape=(nout, nin)).asnumpy()
+ tmp = np.random.uniform(-1.0, 1.0, (nout, nin))
elif self.rand_type == "normal":
- tmp = random.normal(0.0, 1.0, shape=(nout, nin)).asnumpy()
+ tmp = np.random.normal(0.0, 1.0, (nout, nin))
u, _, v = np.linalg.svd(tmp, full_matrices=False) # pylint: disable=invalid-name
if u.shape == tmp.shape:
res = u
diff --git a/python/mxnet/io.py b/python/mxnet/io.py
index 2bace6f526f..201414e8f6e 100644
--- a/python/mxnet/io.py
+++ b/python/mxnet/io.py
@@ -39,8 +39,6 @@
from .ndarray import _ndarray_cls
from .ndarray import array
from .ndarray import concatenate
-from .ndarray import arange
-from .ndarray.random import shuffle as random_shuffle
class DataDesc(namedtuple('DataDesc', ['name', 'shape'])):
"""DataDesc is used to store name, shape, type and layout
@@ -653,14 +651,12 @@ def __init__(self, data, label=None, batch_size=1, shuffle=False,
raise NotImplementedError("`NDArrayIter` only supports ``CSRNDArray``" \
" with `last_batch_handle` set to `discard`.")
+ self.idx = np.arange(self.data[0][1].shape[0])
# shuffle data
if shuffle:
- tmp_idx = arange(self.data[0][1].shape[0], dtype=np.int32)
- self.idx = random_shuffle(tmp_idx, out=tmp_idx).asnumpy()
+ np.random.shuffle(self.idx)
self.data = _shuffle(self.data, self.idx)
self.label = _shuffle(self.label, self.idx)
- else:
- self.idx = np.arange(self.data[0][1].shape[0])
# batching
if last_batch_handle == 'discard':
diff --git a/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ImageClassifier.scala b/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ImageClassifier.scala
index f05b2e2cd8f..45c4e767cb3 100644
--- a/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ImageClassifier.scala
+++ b/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ImageClassifier.scala
@@ -17,7 +17,7 @@
package ml.dmlc.mxnet.infer
-import ml.dmlc.mxnet.{Context, DataDesc, NDArray, Shape}
+import ml.dmlc.mxnet.{DataDesc, NDArray, Shape}
import scala.collection.mutable.ListBuffer
@@ -37,15 +37,13 @@ import javax.imageio.ImageIO
* file://model-dir/synset.txt
* @param inputDescriptors Descriptors defining the input node names, shape,
* layout and Type parameters
- * @param contexts Device Contexts on which you want to run Inference, defaults to CPU.
- * @param epoch Model epoch to load, defaults to 0.
*/
class ImageClassifier(modelPathPrefix: String,
- inputDescriptors: IndexedSeq[DataDesc],
- contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0))
+ inputDescriptors: IndexedSeq[DataDesc])
extends Classifier(modelPathPrefix,
- inputDescriptors, contexts, epoch) {
+ inputDescriptors) {
+
+ val classifier: Classifier = getClassifier(modelPathPrefix, inputDescriptors)
protected[infer] val inputLayout = inputDescriptors.head.layout
@@ -110,11 +108,8 @@ class ImageClassifier(modelPathPrefix: String,
result
}
- private[infer] def getClassifier(modelPathPrefix: String,
- inputDescriptors: IndexedSeq[DataDesc],
- contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0)): Classifier = {
- new Classifier(modelPathPrefix, inputDescriptors, contexts, epoch)
+ def getClassifier(modelPathPrefix: String, inputDescriptors: IndexedSeq[DataDesc]): Classifier = {
+ new Classifier(modelPathPrefix, inputDescriptors)
}
}
diff --git a/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ObjectDetector.scala b/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ObjectDetector.scala
index 5af3fe99a8e..2d83caf2386 100644
--- a/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ObjectDetector.scala
+++ b/scala-package/infer/src/main/scala/ml/dmlc/mxnet/infer/ObjectDetector.scala
@@ -16,14 +16,12 @@
*/
package ml.dmlc.mxnet.infer
-
// scalastyle:off
import java.awt.image.BufferedImage
// scalastyle:on
-
-import ml.dmlc.mxnet.{Context, DataDesc, NDArray}
+import ml.dmlc.mxnet.NDArray
+import ml.dmlc.mxnet.DataDesc
import scala.collection.mutable.ListBuffer
-
/**
* A class for object detection tasks
*
@@ -34,34 +32,29 @@ import scala.collection.mutable.ListBuffer
* file://model-dir/synset.txt
* @param inputDescriptors Descriptors defining the input node names, shape,
* layout and Type parameters
- * @param contexts Device Contexts on which you want to run Inference, defaults to CPU.
- * @param epoch Model epoch to load, defaults to 0.
*/
class ObjectDetector(modelPathPrefix: String,
- inputDescriptors: IndexedSeq[DataDesc],
- contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0)) {
+ inputDescriptors: IndexedSeq[DataDesc]) {
- protected[infer] val imgClassifier: ImageClassifier =
- getImageClassifier(modelPathPrefix, inputDescriptors, contexts, epoch)
+ val imgClassifier: ImageClassifier = getImageClassifier(modelPathPrefix, inputDescriptors)
- protected[infer] val inputShape = imgClassifier.inputShape
+ val inputShape = imgClassifier.inputShape
- protected[infer] val handler = imgClassifier.handler
+ val handler = imgClassifier.handler
- protected[infer] val predictor = imgClassifier.predictor
+ val predictor = imgClassifier.predictor
- protected[infer] val synset = imgClassifier.synset
+ val synset = imgClassifier.synset
- protected[infer] val height = imgClassifier.height
+ val height = imgClassifier.height
- protected[infer] val width = imgClassifier.width
+ val width = imgClassifier.width
/**
* To Detect bounding boxes and corresponding labels
*
* @param inputImage : PathPrefix of the input image
- * @param topK : Get top k elements with maximum probability
+ * @param topK : Get top k elements with maximum probability
* @return List of List of tuples of (class, [probability, xmin, ymin, xmax, ymax])
*/
def imageObjectDetect(inputImage: BufferedImage,
@@ -78,10 +71,9 @@ class ObjectDetector(modelPathPrefix: String,
/**
* Takes input images as NDArrays. Useful when you want to perform multiple operations on
* the input Array, or when you want to pass a batch of input images.
- *
* @param input : Indexed Sequence of NDArrays
- * @param topK : (Optional) How many top_k(sorting will be based on the last axis)
- * elements to return. If not passed, returns all unsorted output.
+ * @param topK : (Optional) How many top_k(sorting will be based on the last axis)
+ * elements to return. If not passed, returns all unsorted output.
* @return List of List of tuples of (class, [probability, xmin, ymin, xmax, ymax])
*/
def objectDetectWithNDArray(input: IndexedSeq[NDArray], topK: Option[Int])
@@ -98,10 +90,10 @@ class ObjectDetector(modelPathPrefix: String,
batchResult.toIndexedSeq
}
- private[infer] def sortAndReformat(predictResultND: NDArray, topK: Option[Int])
+ private def sortAndReformat(predictResultND : NDArray, topK: Option[Int])
: IndexedSeq[(String, Array[Float])] = {
val predictResult: ListBuffer[Array[Float]] = ListBuffer[Array[Float]]()
- val accuracy: ListBuffer[Float] = ListBuffer[Float]()
+ val accuracy : ListBuffer[Float] = ListBuffer[Float]()
// iterating over the all the predictions
val length = predictResultND.shape(0)
@@ -118,7 +110,7 @@ class ObjectDetector(modelPathPrefix: String,
handler.execute(r.dispose())
}
var result = IndexedSeq[(String, Array[Float])]()
- if (topK.isDefined) {
+ if(topK.isDefined) {
var sortedIndices = accuracy.zipWithIndex.sortBy(-_._1).map(_._2)
sortedIndices = sortedIndices.take(topK.get)
// takeRight(5) would provide the output as Array[Accuracy, Xmin, Ymin, Xmax, Ymax
@@ -135,9 +127,8 @@ class ObjectDetector(modelPathPrefix: String,
/**
* To classify batch of input images according to the provided model
- *
* @param inputBatch Input batch of Buffered images
- * @param topK Get top k elements with maximum probability
+ * @param topK Get top k elements with maximum probability
* @return List of list of tuples of (class, probability)
*/
def imageBatchObjectDetect(inputBatch: Traversable[BufferedImage], topK: Option[Int] = None):
@@ -157,12 +148,9 @@ class ObjectDetector(modelPathPrefix: String,
result
}
- private[infer] def getImageClassifier(modelPathPrefix: String,
- inputDescriptors: IndexedSeq[DataDesc],
- contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0)):
+ def getImageClassifier(modelPathPrefix: String, inputDescriptors: IndexedSeq[DataDesc]):
ImageClassifier = {
- new ImageClassifier(modelPathPrefix, inputDescriptors, contexts, epoch)
+ new ImageClassifier(modelPathPrefix, inputDescriptors)
}
}
diff --git a/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ImageClassifierSuite.scala b/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ImageClassifierSuite.scala
index 85059be43aa..96fc80014cc 100644
--- a/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ImageClassifierSuite.scala
+++ b/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ImageClassifierSuite.scala
@@ -17,10 +17,11 @@
package ml.dmlc.mxnet.infer
-import ml.dmlc.mxnet._
+import ml.dmlc.mxnet.{DType, DataDesc, Shape, NDArray}
+
import org.mockito.Matchers._
import org.mockito.Mockito
-import org.scalatest.BeforeAndAfterAll
+import org.scalatest.{BeforeAndAfterAll}
// scalastyle:off
import java.awt.image.BufferedImage
@@ -32,7 +33,7 @@ import java.awt.image.BufferedImage
class ImageClassifierSuite extends ClassifierSuite with BeforeAndAfterAll {
class MyImageClassifier(modelPathPrefix: String,
- inputDescriptors: IndexedSeq[DataDesc])
+ inputDescriptors: IndexedSeq[DataDesc])
extends ImageClassifier(modelPathPrefix, inputDescriptors) {
override def getPredictor(): MyClassyPredictor = {
@@ -40,8 +41,7 @@ class ImageClassifierSuite extends ClassifierSuite with BeforeAndAfterAll {
}
override def getClassifier(modelPathPrefix: String, inputDescriptors:
- IndexedSeq[DataDesc], contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0)): Classifier = {
+ IndexedSeq[DataDesc]): Classifier = {
Mockito.mock(classOf[Classifier])
}
@@ -84,7 +84,7 @@ class ImageClassifierSuite extends ClassifierSuite with BeforeAndAfterAll {
val synset = testImageClassifier.synset
- val predictExpectedOp: List[(String, Float)] =
+ val predictExpectedOp : List[(String, Float)] =
List[(String, Float)]((synset(1), .98f), (synset(2), .97f),
(synset(3), .96f), (synset(0), .99f))
@@ -93,14 +93,13 @@ class ImageClassifierSuite extends ClassifierSuite with BeforeAndAfterAll {
Mockito.doReturn(IndexedSeq(predictExpectedND)).when(testImageClassifier.predictor)
.predictWithNDArray(any(classOf[IndexedSeq[NDArray]]))
- Mockito.doReturn(IndexedSeq(predictExpectedOp))
- .when(testImageClassifier.getClassifier(modelPath, inputDescriptor))
+ Mockito.doReturn(IndexedSeq(predictExpectedOp)).when(testImageClassifier.classifier)
.classifyWithNDArray(any(classOf[IndexedSeq[NDArray]]), Some(anyInt()))
val predictResult: IndexedSeq[IndexedSeq[(String, Float)]] =
testImageClassifier.classifyImage(inputImage, Some(4))
- for (i <- predictExpected.indices) {
+ for(i <- predictExpected.indices) {
assertResult(predictExpected(i).sortBy(-_)) {
predictResult(i).map(_._2).toArray
}
@@ -120,15 +119,15 @@ class ImageClassifierSuite extends ClassifierSuite with BeforeAndAfterAll {
val predictExpected: IndexedSeq[Array[Array[Float]]] =
IndexedSeq[Array[Array[Float]]](Array(Array(.98f, 0.97f, 0.96f, 0.99f),
- Array(.98f, 0.97f, 0.96f, 0.99f)))
+ Array(.98f, 0.97f, 0.96f, 0.99f)))
val synset = testImageClassifier.synset
- val predictExpectedOp: List[List[(String, Float)]] =
+ val predictExpectedOp : List[List[(String, Float)]] =
List[List[(String, Float)]](List((synset(1), .98f), (synset(2), .97f),
(synset(3), .96f), (synset(0), .99f)),
List((synset(1), .98f), (synset(2), .97f),
- (synset(3), .96f), (synset(0), .99f)))
+ (synset(3), .96f), (synset(0), .99f)))
val predictExpectedND: NDArray = NDArray.array(predictExpected.flatten.flatten.toArray,
Shape(2, 4))
@@ -136,8 +135,7 @@ class ImageClassifierSuite extends ClassifierSuite with BeforeAndAfterAll {
Mockito.doReturn(IndexedSeq(predictExpectedND)).when(testImageClassifier.predictor)
.predictWithNDArray(any(classOf[IndexedSeq[NDArray]]))
- Mockito.doReturn(IndexedSeq(predictExpectedOp))
- .when(testImageClassifier.getClassifier(modelPath, inputDescriptor))
+ Mockito.doReturn(IndexedSeq(predictExpectedOp)).when(testImageClassifier.classifier)
.classifyWithNDArray(any(classOf[IndexedSeq[NDArray]]), Some(anyInt()))
val result: IndexedSeq[IndexedSeq[(String, Float)]] =
diff --git a/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ObjectDetectorSuite.scala b/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ObjectDetectorSuite.scala
index 5e6f32f1107..a691aa36ecb 100644
--- a/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ObjectDetectorSuite.scala
+++ b/scala-package/infer/src/test/scala/ml/dmlc/mxnet/infer/ObjectDetectorSuite.scala
@@ -23,7 +23,7 @@ import java.awt.image.BufferedImage
// scalastyle:on
import ml.dmlc.mxnet.Context
import ml.dmlc.mxnet.DataDesc
-import ml.dmlc.mxnet.{Context, NDArray, Shape}
+import ml.dmlc.mxnet.{NDArray, Shape}
import org.mockito.Matchers.any
import org.mockito.Mockito
import org.scalatest.BeforeAndAfterAll
@@ -36,8 +36,7 @@ class ObjectDetectorSuite extends ClassifierSuite with BeforeAndAfterAll {
extends ObjectDetector(modelPathPrefix, inputDescriptors) {
override def getImageClassifier(modelPathPrefix: String, inputDescriptors:
- IndexedSeq[DataDesc], contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0)): ImageClassifier = {
+ IndexedSeq[DataDesc]): ImageClassifier = {
new MyImageClassifier(modelPathPrefix, inputDescriptors)
}
@@ -45,15 +44,13 @@ class ObjectDetectorSuite extends ClassifierSuite with BeforeAndAfterAll {
class MyImageClassifier(modelPathPrefix: String,
protected override val inputDescriptors: IndexedSeq[DataDesc])
- extends ImageClassifier(modelPathPrefix, inputDescriptors, Context.cpu(), Some(0)) {
+ extends ImageClassifier(modelPathPrefix, inputDescriptors) {
override def getPredictor(): MyClassyPredictor = {
Mockito.mock(classOf[MyClassyPredictor])
}
- override def getClassifier(modelPathPrefix: String, inputDescriptors: IndexedSeq[DataDesc],
- contexts: Array[Context] = Context.cpu(),
- epoch: Option[Int] = Some(0)):
+ override def getClassifier(modelPathPrefix: String, inputDescriptors: IndexedSeq[DataDesc]):
Classifier = {
new MyClassifier(modelPathPrefix, inputDescriptors)
}
diff --git a/scala-package/native/osx-x86_64-cpu/pom.xml b/scala-package/native/osx-x86_64-cpu/pom.xml
index 529b1264b51..fa0f5b6c49d 100644
--- a/scala-package/native/osx-x86_64-cpu/pom.xml
+++ b/scala-package/native/osx-x86_64-cpu/pom.xml
@@ -67,7 +67,7 @@
<linkerMiddleOption>-Wl,-x</linkerMiddleOption>
<linkerMiddleOption>${lddeps}</linkerMiddleOption>
<linkerMiddleOption>-force_load ../../../lib/libmxnet.a</linkerMiddleOption>
- <linkerMiddleOption>-force_load ../../../3rdparty/nnvm/lib/libnnvm.a</linkerMiddleOption>
+ <linkerMiddleOption>-force_load ../../../nnvm/lib/libnnvm.a</linkerMiddleOption>
</linkerMiddleOptions>
<linkerEndOptions>
<linkerEndOption>${ldflags}</linkerEndOption>
diff --git a/setup-utils/install-mxnet-virtualenv.sh b/setup-utils/install-mxnet-virtualenv.sh
deleted file mode 100755
index 5e00f79647e..00000000000
--- a/setup-utils/install-mxnet-virtualenv.sh
+++ /dev/null
@@ -1,123 +0,0 @@
-#!/usr/bin/env bash
-
-# Licensed to the Apache Software Foundation (ASF) under one
-# or more contributor license agreements. See the NOTICE file
-# distributed with this work for additional information
-# regarding copyright ownership. The ASF licenses this file
-# to you under the Apache License, Version 2.0 (the
-# "License"); you may not use this file except in compliance
-# with the License. You may obtain a copy of the License at
-#
-# http://www.apache.org/licenses/LICENSE-2.0
-#
-# Unless required by applicable law or agreed to in writing,
-# software distributed under the License is distributed on an
-# "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY
-# KIND, either express or implied. See the License for the
-# specific language governing permissions and limitations
-# under the License.
-
-######################################################################
-# This script installs MXNet for Python in a virtualenv on OSX and ubuntu
-######################################################################
-set -e
-#set -x
-
-BUILDIR=build
-VENV=mxnet_py3
-
-setup_virtualenv() {
- if [ ! -d $VENV ];then
- virtualenv -p `which python3` $VENV
- fi
- source $VENV/bin/activate
-}
-
-gpu_count() {
- nvidia-smi -L | wc -l
-}
-
-detect_platform() {
- unameOut="$(uname -s)"
- case "${unameOut}" in
- Linux*)
- distro=$(awk -F= '/^NAME/{gsub(/"/, "", $2); print $2}' /etc/os-release)
- machine="Linux/$distro"
- ;;
- Darwin*) machine=Mac;;
- CYGWIN*) machine=Cygwin;;
- MINGW*) machine=MinGw;;
- *) machine="UNKNOWN:${unameOut}"
- esac
- echo ${machine}
-}
-
-
-if [ $(gpu_count) -ge 1 ];then
- USE_CUDA=ON
-else
- USE_CUDA=OFF
-fi
-
-PLATFORM=$(detect_platform)
-echo "Detected platform '$PLATFORM'"
-
-if [ $PLATFORM = "Mac" ];then
- USE_OPENMP=OFF
-else
- USE_OPENMP=ON
-fi
-
-if [ $PLATFORM = "Linux/Ubuntu" ];then
- install_dependencies_ubuntu() {
- sudo apt-get update
- sudo apt-get install -y build-essential libatlas-base-dev libopencv-dev graphviz virtualenv cmake\
- ninja-build libopenblas-dev liblapack-dev python3 python3-dev
- }
- echo "Installing build dependencies in Ubuntu!"
- install_dependencies_ubuntu
-fi
-
-echo "Preparing a Python virtualenv in ${VENV}"
-setup_virtualenv
-
-echo "Building MXNet core. This can take a few minutes..."
-build_mxnet() {
- pushd .
- set -x
- mkdir -p $BUILDIR && cd $BUILDIR
- cmake -DUSE_CUDA=$USE_CUDA -DUSE_OPENCV=ON -DUSE_OPENMP=$USE_OPENMP -DUSE_SIGNAL_HANDLER=ON -DCMAKE_BUILD_TYPE=Release -GNinja ..
- ninja
- set +x
- popd
-}
-
-
-build_mxnet
-
-echo "Installing mxnet under virtualenv ${VENV}"
-install_mxnet() {
- pushd .
- cd python
- pip3 install -e .
- pip3 install opencv-python matplotlib graphviz jupyter ipython
- popd
-}
-
-install_mxnet
-
-echo "
-
-========================================================================================
-Done! MXNet for Python installation is complete. Go ahead and explore MXNet with Python.
-========================================================================================
-
-Use the following command to enter the virtualenv:
-$ source ${VENV}/bin/activate
-$ iptyhon
-
-You can then start using mxnet
-
-import mxnet as mx
-x = mx.nd.ones((5,5))
-"
diff --git a/src/engine/threaded_engine.cc b/src/engine/threaded_engine.cc
index ca5602bb482..77b14b43a7b 100644
--- a/src/engine/threaded_engine.cc
+++ b/src/engine/threaded_engine.cc
@@ -326,7 +326,8 @@ void ThreadedEngine::PushSync(SyncFn exec_fn, Context exec_ctx,
FnProperty prop,
int priority,
const char* opr_name) {
- if (!bulk_size() || prop != FnProperty::kNormal || priority) {
+ BulkStatus& bulk_status = *BulkStatusStore::Get();
+ if (!bulk_status.bulk_size || prop != FnProperty::kNormal || priority) {
this->PushAsync([exec_fn](RunContext ctx, CallbackOnComplete on_complete) {
exec_fn(ctx);
on_complete();
@@ -334,9 +335,9 @@ void ThreadedEngine::PushSync(SyncFn exec_fn, Context exec_ctx,
return;
}
- const BulkStatus& bulk_status = *BulkStatusStore::Get();
if (bulk_status.count && exec_ctx != bulk_status.ctx) BulkFlush();
BulkAppend(exec_fn, exec_ctx, const_vars, mutable_vars);
+ return;
}
void ThreadedEngine::DeleteVariable(SyncFn delete_fn,
diff --git a/src/engine/threaded_engine.h b/src/engine/threaded_engine.h
index d72784d0498..1b9453f903b 100644
--- a/src/engine/threaded_engine.h
+++ b/src/engine/threaded_engine.h
@@ -398,7 +398,7 @@ class ThreadedEngine : public Engine {
}
int bulk_size() const override {
- return profiler::Profiler::Get()->AggregateRunning() ? 0 : BulkStatusStore::Get()->bulk_size;
+ return BulkStatusStore::Get()->bulk_size;
}
int set_bulk_size(int bulk_size) override {
diff --git a/src/executor/graph_executor.cc b/src/executor/graph_executor.cc
index fa5931e5c84..ee97649768b 100644
--- a/src/executor/graph_executor.cc
+++ b/src/executor/graph_executor.cc
@@ -1348,8 +1348,7 @@ void GraphExecutor::InitOpSegs() {
// Generate segments based on the graph structure
bool prefer_bulk_exec_inference = dmlc::GetEnv("MXNET_EXEC_BULK_EXEC_INFERENCE", true);
// Whether to perform bulk exec for training
- bool prefer_bulk_exec = dmlc::GetEnv("MXNET_EXEC_BULK_EXEC_TRAIN", 1)
- && !profiler::Profiler::Get()->AggregateEnabled();
+ bool prefer_bulk_exec = dmlc::GetEnv("MXNET_EXEC_BULK_EXEC_TRAIN", 1);
bool is_training = num_forward_nodes_ != total_num_nodes;
@@ -1360,6 +1359,8 @@ void GraphExecutor::InitOpSegs() {
if (prefer_bulk_exec_inference && !is_training) {
this->BulkInferenceOpSegs();
}
+
+ return;
}
void GraphExecutor::BulkTrainingOpSegs(size_t total_num_nodes) {
diff --git a/src/io/iter_sparse_batchloader.h b/src/io/iter_sparse_batchloader.h
index 398d6e00fe7..d5c9bd2f457 100644
--- a/src/io/iter_sparse_batchloader.h
+++ b/src/io/iter_sparse_batchloader.h
@@ -68,36 +68,53 @@ class SparseBatchLoader : public BatchLoader, public SparseIIterator<TBlobBatch>
// if overflown from previous round, directly return false, until before first is called
if (num_overflow_ != 0) return false;
index_t top = 0;
- offsets_.clear();
+ inst_cache_.clear();
while (sparse_base_->Next()) {
- const DataInst& inst = sparse_base_->Value();
- // initialize the data buffer, only called once
- if (data_.size() == 0) this->InitData(inst);
- // initialize the number of elements in each buffer, called once per batch
- if (offsets_.size() == 0) offsets_.resize(inst.data.size(), 0);
- CopyData(inst, top);
- if (++top >= param_.batch_size) {
- SetOutputShape();
- return true;
- }
+ inst_cache_.emplace_back(sparse_base_->Value());
+ if (inst_cache_.size() >= param_.batch_size) break;
}
- if (top != 0) {
- CHECK_NE(param_.round_batch, 0)
- << "round_batch = False is not supported for sparse data iterator";
+ // no more data instance
+ if (inst_cache_.size() == 0) {
+ return false;
+ }
+ if (inst_cache_.size() < param_.batch_size) {
+ CHECK_GT(param_.round_batch, 0);
num_overflow_ = 0;
sparse_base_->BeforeFirst();
- for (; top < param_.batch_size; ++top, ++num_overflow_) {
+ for (; inst_cache_.size() < param_.batch_size; ++num_overflow_) {
CHECK(sparse_base_->Next()) << "number of input must be bigger than batch size";
- const DataInst& inst = sparse_base_->Value();
- // copy data
- CopyData(inst, top);
+ inst_cache_.emplace_back(sparse_base_->Value());
}
- SetOutputShape();
- out_.num_batch_padd = num_overflow_;
- return true;
}
- // no more data instance
- return false;
+ out_.num_batch_padd = num_overflow_;
+ CHECK_EQ(inst_cache_.size(), param_.batch_size);
+ this->InitDataFromBatch();
+ for (size_t j = 0; j < inst_cache_.size(); j++) {
+ const auto& d = inst_cache_[j];
+ out_.inst_index[top] = d.index;
+ // TODO(haibin) double check the type?
+ int64_t unit_size = 0;
+ for (size_t i = 0; i < d.data.size(); ++i) {
+ // indptr tensor
+ if (IsIndPtr(i)) {
+ auto indptr = data_[i].get<cpu, 1, int64_t>();
+ if (j == 0) indptr[0] = 0;
+ indptr[j + 1] = indptr[j] + unit_size;
+ offsets_[i] = j;
+ } else {
+ // indices and values tensor
+ unit_size = d.data[i].shape_.Size();
+ MSHADOW_TYPE_SWITCH(data_[i].type_flag_, DType, {
+ const auto begin = offsets_[i];
+ const auto end = offsets_[i] + unit_size;
+ mshadow::Copy(data_[i].get<cpu, 1, DType>().Slice(begin, end),
+ d.data[i].get_with_shape<cpu, 1, DType>(mshadow::Shape1(unit_size)));
+ });
+ offsets_[i] += unit_size;
+ }
+ }
+ }
+ return true;
}
virtual const TBlobBatch &Value(void) const {
@@ -121,16 +138,14 @@ class SparseBatchLoader : public BatchLoader, public SparseIIterator<TBlobBatch>
private:
/*! \brief base sparse iterator */
SparseIIterator<DataInst> *sparse_base_;
+ /*! \brief data instances */
+ std::vector<DataInst> inst_cache_;
/*! \brief data storage type */
NDArrayStorageType data_stype_;
/*! \brief data label type */
NDArrayStorageType label_stype_;
- /*! \brief tensor offsets for slicing */
+ /*! \brief tensor offset for slicing */
std::vector<size_t> offsets_;
- /*! \brief tensor dtypes */
- std::vector<int> dtypes_;
- /*! \brief whether the offset correspond to an indptr array */
- std::vector<bool> indptr_;
// check whether ith position is the indptr tensor for a CSR tensor
inline bool IsIndPtr(size_t i) {
@@ -142,109 +157,44 @@ class SparseBatchLoader : public BatchLoader, public SparseIIterator<TBlobBatch>
return true;
}
// label indptr
- if (i == label_indptr_offset && label_stype_ == kCSRStorage &&
- data_stype_ == kCSRStorage) {
+ if (i == label_indptr_offset && label_stype_ == kCSRStorage && data_stype_ == kCSRStorage) {
return true;
}
return false;
}
// initialize the data holder by using from the batch
- inline void InitData(const DataInst& first_inst) {
+ inline void InitDataFromBatch() {
CHECK(data_stype_ == kCSRStorage || label_stype_ == kCSRStorage);
+ CHECK_GT(inst_cache_.size(), 0);
out_.data.clear();
data_.clear();
offsets_.clear();
- indptr_.clear();
-
- // num_arrays is the number of arrays in inputs
- // if both data and label are in the csr format,
- // num_arrays will be 3 + 3 = 6.
- size_t num_arrays = first_inst.data.size();
- data_.resize(num_arrays);
- offsets_.resize(num_arrays, 0);
- indptr_.resize(num_arrays, false);
- // tensor buffer sizes
- std::vector<size_t> buff_sizes(num_arrays, 0);
- dtypes_.resize(num_arrays);
- out_.data.resize(num_arrays);
- // estimate the memory required for a batch
- for (size_t i = 0; i < num_arrays; ++i) {
- // shape for indptr
+
+ size_t total_size = inst_cache_[0].data.size();
+ data_.resize(total_size);
+ offsets_.resize(total_size, 0);
+ std::vector<size_t> vec_sizes(total_size, 0);
+ // accumulate the memory required for a batch
+ for (size_t i = 0; i < total_size; ++i) {
+ size_t size = 0;
+ // vec_size for indptr
if (IsIndPtr(i)) {
- buff_sizes[i] = param_.batch_size + 1;
- indptr_[i] = true;
+ size = param_.batch_size + 1;
} else {
- // estimated the size for the whole batch based on the first instance
- buff_sizes[i] = first_inst.data[i].Size() * param_.batch_size;
- indptr_[i] = false;
+ for (const auto &d : inst_cache_) size += d.data[i].shape_.Size();
}
- dtypes_[i] = first_inst.data[i].type_flag_;
+ vec_sizes[i] = size;
}
- CHECK_EQ(buff_sizes[0], buff_sizes[1]);
- // allocate buffer
- for (size_t i = 0; i < num_arrays; ++i) {
+ CHECK_EQ(vec_sizes[0], vec_sizes[1]);
+ for (size_t i = 0; i < total_size; ++i) {
+ int src_type_flag = inst_cache_[0].data[i].type_flag_;
// init object attributes
- TShape dst_shape(mshadow::Shape1(buff_sizes[i]));
- data_[i].resize(mshadow::Shape1(buff_sizes[i]), dtypes_[i]);
+ TShape dst_shape(mshadow::Shape1(vec_sizes[i]));
+ data_[i].resize(mshadow::Shape1(vec_sizes[i]), src_type_flag);
CHECK(data_[i].dptr_ != nullptr);
- }
- }
-
- /* \brief set the shape of the outputs based on actual shapes */
- inline void SetOutputShape() {
- for (size_t i = 0; i < out_.data.size(); i++) {
- out_.data[i] = TBlob(data_[i].dptr_, mshadow::Shape1(offsets_[i]),
- Context::kCPU, dtypes_[i]);
- }
- }
-
- /* \brief increase the size of i-th data buffer by a factor of 2, while retaining the content */
- inline void ResizeBuffer(size_t src_size, size_t i) {
- MSHADOW_TYPE_SWITCH(data_[i].type_flag_, DType, {
- TBlobContainer temp;
- temp.resize(mshadow::Shape1(src_size), dtypes_[i]);
- mshadow::Copy(temp.get<cpu, 1, DType>(), data_[i].get<cpu, 1, DType>().Slice(0, src_size));
- // increase the size of space exponentially
- size_t capacity = data_[i].Size();
- capacity = capacity * 2 + 1;
- data_[i] = TBlobContainer();
- data_[i].resize(mshadow::Shape1(capacity), dtypes_[i]);
- // copy back
- mshadow::Copy(data_[i].get<cpu, 1, DType>().Slice(0, src_size), temp.get<cpu, 1, DType>());
- });
- }
-
- /* \brief copy the data instance to data buffer */
- void CopyData(const DataInst& inst, const size_t top) {
- int64_t unit_size = 0;
- out_.inst_index[top] = inst.index;
- for (size_t i = 0; i < inst.data.size(); ++i) {
- if (!indptr_[i]) {
- // indices and values tensor
- unit_size = inst.data[i].shape_.Size();
- MSHADOW_TYPE_SWITCH(data_[i].type_flag_, DType, {
- const size_t begin = offsets_[i];
- const size_t end = offsets_[i] + unit_size;
- size_t capacity = data_[i].Size();
- // resize the data buffer if estimated space is not sufficient
- while (capacity < end) {
- ResizeBuffer(begin, i);
- capacity = data_[i].Size();
- }
- mshadow::Copy(data_[i].get<cpu, 1, DType>().Slice(begin, end),
- inst.data[i].get_with_shape<cpu, 1, DType>(mshadow::Shape1(unit_size)));
- });
- offsets_[i] += unit_size;
- } else {
- // indptr placeholder
- auto indptr = data_[i].get<cpu, 1, int64_t>();
- // initialize the first indptr, which is always 0
- if (top == 0) indptr[0] = 0;
- indptr[top + 1] = indptr[top] + unit_size;
- offsets_[i] = top + 2;
- }
+ out_.data.push_back(TBlob(data_[i].dptr_, dst_shape, cpu::kDevMask, src_type_flag));
}
}
}; // class BatchLoader
diff --git a/src/ndarray/ndarray.cc b/src/ndarray/ndarray.cc
index 52b96fad692..35dd4453576 100644
--- a/src/ndarray/ndarray.cc
+++ b/src/ndarray/ndarray.cc
@@ -742,8 +742,10 @@ void NDArray::SetTBlob() const {
auto stype = storage_type();
if (stype == kDefaultStorage) {
#if MXNET_USE_MKLDNN == 1
- CHECK(!IsMKLDNNData()) << "We can't generate TBlob for MKLDNN data. "
- << "Please use Reorder2Default() to generate a new NDArray first";
+ if (IsMKLDNNData()) {
+ ptr_->Reorder2Default();
+ dptr = static_cast<char*>(ptr_->shandle.dptr);
+ }
#endif
dptr += byte_offset_;
} else if (stype == kCSRStorage || stype == kRowSparseStorage) {
diff --git a/src/operator/contrib/quadratic_op-inl.h b/src/operator/contrib/quadratic_op-inl.h
index 71cb76a7b56..8d73a4286f6 100644
--- a/src/operator/contrib/quadratic_op-inl.h
+++ b/src/operator/contrib/quadratic_op-inl.h
@@ -32,7 +32,6 @@
#include "../mxnet_op.h"
#include "../operator_common.h"
#include "../elemwise_op_common.h"
-#include "../tensor/init_op.h"
namespace mxnet {
namespace op {
@@ -74,33 +73,6 @@ inline bool QuadraticOpType(const nnvm::NodeAttrs& attrs,
return out_attrs->at(0) != -1;
}
-inline bool QuadraticOpStorageType(const nnvm::NodeAttrs& attrs,
- const int dev_mask,
- DispatchMode* dispatch_mode,
- std::vector<int>* in_attrs,
- std::vector<int>* out_attrs) {
- CHECK_EQ(in_attrs->size(), 1U);
- CHECK_EQ(out_attrs->size(), 1U);
- const QuadraticParam& param = nnvm::get<QuadraticParam>(attrs.parsed);
- const int in_stype = in_attrs->at(0);
- int& out_stype = out_attrs->at(0);
- bool dispatched = false;
- if (!dispatched && in_stype == kDefaultStorage) {
- // dns -> dns
- dispatched = storage_type_assign(&out_stype, kDefaultStorage,
- dispatch_mode, DispatchMode::kFCompute);
- }
- if (!dispatched && in_stype == kCSRStorage && param.c == 0.0) {
- // csr -> csr
- dispatched = storage_type_assign(&out_stype, kCSRStorage,
- dispatch_mode, DispatchMode::kFComputeEx);
- }
- if (!dispatched) {
- dispatched = dispatch_fallback(out_attrs, dispatch_mode);
- }
- return dispatched;
-}
-
template<int req>
struct quadratic_forward {
template<typename DType>
@@ -142,61 +114,6 @@ void QuadraticOpForward(const nnvm::NodeAttrs& attrs,
});
}
-template<typename xpu>
-void QuadraticOpForwardCsrImpl(const QuadraticParam& param,
- const OpContext& ctx,
- const NDArray& input,
- const OpReqType req,
- const NDArray& output) {
- using namespace mshadow;
- using namespace mxnet_op;
- using namespace csr;
- if (req == kNullOp) return;
- CHECK_EQ(req, kWriteTo) << "QuadraticOp with CSR only supports kWriteTo";
- Stream<xpu> *s = ctx.get_stream<xpu>();
- if (!input.storage_initialized()) {
- FillZerosCsrImpl(s, output);
- return;
- }
- const nnvm::dim_t nnz = input.storage_shape()[0];
- const nnvm::dim_t num_rows = output.shape()[0];
- output.CheckAndAlloc({Shape1(num_rows + 1), Shape1(nnz)});
- CHECK_EQ(output.aux_type(kIdx), output.aux_type(kIndPtr))
- << "The dtypes of indices and indptr don't match";
- MSHADOW_TYPE_SWITCH(output.dtype(), DType, {
- MSHADOW_IDX_TYPE_SWITCH(output.aux_type(kIdx), IType, {
- MXNET_ASSIGN_REQ_SWITCH(req, req_type, {
- Kernel<quadratic_forward<req_type>, xpu>::Launch(
- s, nnz, output.data().dptr<DType>(), input.data().dptr<DType>(),
- param.a, param.b, param.c);
- Copy(output.aux_data(kIdx).FlatTo1D<xpu, IType>(s),
- input.aux_data(kIdx).FlatTo1D<xpu, IType>(s), s);
- Copy(output.aux_data(kIndPtr).FlatTo1D<xpu, IType>(s),
- input.aux_data(kIndPtr).FlatTo1D<xpu, IType>(s), s);
- });
- });
- });
-}
-
-template<typename xpu>
-void QuadraticOpForwardEx(const nnvm::NodeAttrs& attrs,
- const OpContext& ctx,
- const std::vector<NDArray>& inputs,
- const std::vector<OpReqType>& req,
- const std::vector<NDArray>& outputs) {
- CHECK_EQ(inputs.size(), 1U);
- CHECK_EQ(outputs.size(), 1U);
- CHECK_EQ(req.size(), 1U);
- const QuadraticParam& param = nnvm::get<QuadraticParam>(attrs.parsed);
- const auto in_stype = inputs[0].storage_type();
- const auto out_stype = outputs[0].storage_type();
- if (in_stype == kCSRStorage && out_stype == kCSRStorage && param.c == 0.0) {
- QuadraticOpForwardCsrImpl<xpu>(param, ctx, inputs[0], req[0], outputs[0]);
- } else {
- LogUnimplementedOp(attrs, ctx, inputs, req, outputs);
- }
-}
-
template<typename xpu>
void QuadraticOpBackward(const nnvm::NodeAttrs& attrs,
const OpContext& ctx,
diff --git a/src/operator/contrib/quadratic_op.cc b/src/operator/contrib/quadratic_op.cc
index d8b2d785c79..5b2d84cfcba 100644
--- a/src/operator/contrib/quadratic_op.cc
+++ b/src/operator/contrib/quadratic_op.cc
@@ -38,11 +38,6 @@ Example::
x = [[1, 2], [3, 4]]
y = quadratic(data=x, a=1, b=2, c=3)
y = [[6, 11], [18, 27]]
-
-The storage type of ``quadratic`` output depends on storage types of inputs
- - quadratic(csr, a, b, 0) = csr
- - quadratic(default, a, b, c) = default
-
)code" ADD_FILELINE)
.set_attr_parser(ParamParser<QuadraticParam>)
.set_num_inputs(1)
@@ -53,7 +48,6 @@ The storage type of ``quadratic`` output depends on storage types of inputs
})
.set_attr<nnvm::FInferShape>("FInferShape", QuadraticOpShape)
.set_attr<nnvm::FInferType>("FInferType", QuadraticOpType)
-.set_attr<FInferStorageType>("FInferStorageType", QuadraticOpStorageType)
.set_attr<FCompute>("FCompute<cpu>", QuadraticOpForward<cpu>)
.set_attr<nnvm::FGradient>("FGradient", ElemwiseGradUseIn{"_contrib_backward_quadratic"})
.set_attr<nnvm::FInplaceOption>("FInplaceOption",
@@ -68,8 +62,7 @@ NNVM_REGISTER_OP(_contrib_backward_quadratic)
.set_num_inputs(2)
.set_num_outputs(1)
.set_attr<nnvm::TIsBackward>("TIsBackward", true)
-.set_attr<FCompute>("FCompute<cpu>", QuadraticOpBackward<cpu>)
-.set_attr<FComputeEx>("FComputeEx<cpu>", QuadraticOpForwardEx<cpu>);
+.set_attr<FCompute>("FCompute<cpu>", QuadraticOpBackward<cpu>);
} // namespace op
} // namespace mxnet
diff --git a/src/operator/contrib/quadratic_op.cu b/src/operator/contrib/quadratic_op.cu
index 72d15ab3749..ede773a7ea3 100644
--- a/src/operator/contrib/quadratic_op.cu
+++ b/src/operator/contrib/quadratic_op.cu
@@ -27,7 +27,6 @@ namespace mxnet {
namespace op {
NNVM_REGISTER_OP(_contrib_quadratic)
-.set_attr<FComputeEx>("FComputeEx<gpu>", QuadraticOpForwardEx<gpu>)
.set_attr<FCompute>("FCompute<gpu>", QuadraticOpForward<gpu>);
NNVM_REGISTER_OP(_contrib_backward_quadratic)
diff --git a/src/operator/mxnet_op.h b/src/operator/mxnet_op.h
index c3f6dc6558e..30b3c8577ce 100644
--- a/src/operator/mxnet_op.h
+++ b/src/operator/mxnet_op.h
@@ -322,7 +322,7 @@ MSHADOW_CINLINE void copy(mshadow::Stream<xpu> *s, const TBlob& to, const TBlob&
CHECK_EQ(from.dev_mask(), to.dev_mask());
MSHADOW_TYPE_SWITCH(to.type_flag_, DType, {
if (to.type_flag_ == from.type_flag_) {
- mshadow::Copy(to.FlatTo1D<xpu, DType>(s), from.FlatTo1D<xpu, DType>(s), s);
+ mshadow::Copy(to.FlatTo1D<xpu, DType>(), from.FlatTo1D<xpu, DType>(), s);
} else {
MSHADOW_TYPE_SWITCH(from.type_flag_, SrcDType, {
to.FlatTo1D<xpu, DType>(s) = mshadow::expr::tcast<DType>(from.FlatTo1D<xpu, SrcDType>(s));
diff --git a/src/operator/nn/batch_norm.cu b/src/operator/nn/batch_norm.cu
index 703ed398938..c310a93d700 100644
--- a/src/operator/nn/batch_norm.cu
+++ b/src/operator/nn/batch_norm.cu
@@ -708,6 +708,8 @@ void BatchNormGradCompute<gpu>(const nnvm::NodeAttrs& attrs,
})
}
#else
+ aux_states[batchnorm::kMovingMean] = inputs[6];
+ aux_states[batchnorm::kMovingVar] = inputs[7];
MSHADOW_REAL_TYPE_SWITCH_EX(out_grad[0].type_flag_, DType, AccReal, {
BatchNormBackward<gpu, DType, AccReal>(ctx, param, inputs, req, outputs);
});
diff --git a/src/operator/nn/convolution.cc b/src/operator/nn/convolution.cc
index 7aafe9d82f7..79f2e800ee1 100644
--- a/src/operator/nn/convolution.cc
+++ b/src/operator/nn/convolution.cc
@@ -468,7 +468,7 @@ There are other options to tune the performance.
else
return std::vector<std::string>{"data", "weight", "bias"};
})
-.set_attr<nnvm::FListOutputNames>("FListOutputNames",
+.set_attr<nnvm::FListInputNames>("FListOutputNames",
[](const NodeAttrs& attrs) {
return std::vector<std::string>{"output"};
})
diff --git a/src/operator/nn/fully_connected.cc b/src/operator/nn/fully_connected.cc
index 278c130064f..475b6362516 100644
--- a/src/operator/nn/fully_connected.cc
+++ b/src/operator/nn/fully_connected.cc
@@ -267,7 +267,7 @@ This could be used for model inference with `row_sparse` weights trained with `S
return std::vector<std::string>{"data", "weight"};
}
})
-.set_attr<nnvm::FListOutputNames>("FListOutputNames",
+.set_attr<nnvm::FListInputNames>("FListOutputNames",
[](const NodeAttrs& attrs) {
return std::vector<std::string>{"output"};
})
diff --git a/src/operator/nn/lrn.cc b/src/operator/nn/lrn.cc
index 68d32617e9d..2359b49abab 100644
--- a/src/operator/nn/lrn.cc
+++ b/src/operator/nn/lrn.cc
@@ -181,14 +181,6 @@ number of kernels in the layer.
.set_attr<nnvm::FInferShape>("FInferShape", LRNShape)
.set_attr<nnvm::FInferType>("FInferType", LRNType)
.set_attr<FInferStorageType>("FInferStorageType", LRNForwardInferStorageType)
-.set_attr<nnvm::FListInputNames>("FListInputNames",
- [](const NodeAttrs& attrs) {
- return std::vector<std::string>{"data"};
-})
-.set_attr<nnvm::FListInputNames>("FListOutputNames",
- [](const NodeAttrs& attrs) {
- return std::vector<std::string>{"output", "tmp_norm"};
-})
.set_attr<FCompute>("FCompute<cpu>", LRNCompute<cpu>)
#if MXNET_USE_MKLDNN == 1
.set_attr<FComputeEx>("FComputeEx<cpu>", LRNComputeExCPU)
diff --git a/src/operator/optimizer_op-inl.h b/src/operator/optimizer_op-inl.h
index 104f20a61ee..55d215602ee 100644
--- a/src/operator/optimizer_op-inl.h
+++ b/src/operator/optimizer_op-inl.h
@@ -749,9 +749,6 @@ inline void AdamUpdate(const nnvm::NodeAttrs& attrs,
});
}
-template<int req, typename xpu>
-struct AdamDnsRspDnsKernel;
-
/*!
* Note: this kernel performs sparse adam update. For each row-slice in row_sparse
* gradient, it finds the corresponding elements in weight, mean and var and performs
@@ -759,7 +756,7 @@ struct AdamDnsRspDnsKernel;
* The kernel assumes dense weight/mean/var, and row_sparse gradient
*/
template<int req>
-struct AdamDnsRspDnsKernel<req, cpu> {
+struct AdamDnsRspDnsKernel {
template<typename DType, typename IType>
MSHADOW_XINLINE static void Map(int i, const nnvm::dim_t row_length, DType* out_data,
DType* mean_data, DType* var_data, const DType* weight_data, const IType* grad_idx,
@@ -791,33 +788,6 @@ struct AdamDnsRspDnsKernel<req, cpu> {
};
-template<int req>
-struct AdamDnsRspDnsKernel<req, gpu> {
- template<typename DType, typename IType>
- MSHADOW_XINLINE static void Map(int i, const nnvm::dim_t row_length, DType* out_data,
- DType* mean_data, DType* var_data, const DType* weight_data, const IType* grad_idx,
- const DType* grad_data, const DType clip_gradient, const DType beta1, const DType beta2,
- const DType lr, const DType wd, const DType epsilon, const DType rescale_grad) {
- using nnvm::dim_t;
- using namespace mshadow_op;
- const dim_t row_id = i / row_length;
- const dim_t col_id = i % row_length;
- const dim_t row_offset = grad_idx[row_id] * row_length;
- // index in data/mean/var
- const dim_t data_i = row_offset + col_id;
- // index in grad
- DType grad_rescaled = grad_data[i] * rescale_grad + weight_data[data_i] * wd;
- if (clip_gradient >= 0.0f) {
- grad_rescaled = clip::Map(grad_rescaled, clip_gradient);
- }
- mean_data[data_i] = beta1 * mean_data[data_i] + (1.f - beta1) * grad_rescaled;
- var_data[data_i] = beta2 * var_data[data_i] +
- (1.f - beta2) * grad_rescaled * grad_rescaled;
- KERNEL_ASSIGN(out_data[data_i], req, weight_data[data_i] - lr * mean_data[data_i] /
- (square_root::Map(var_data[data_i]) + epsilon));
- }
-};
-
template<typename xpu>
inline void AdamUpdateDnsRspDnsImpl(const AdamParam& param,
const OpContext& ctx,
@@ -847,12 +817,8 @@ inline void AdamUpdateDnsRspDnsImpl(const AdamParam& param,
DType* out_data = out->dptr<DType>();
nnvm::dim_t num_rows = grad.aux_shape(kIdx)[0];
const auto row_length = weight.shape_.ProdShape(1, weight.ndim());
- size_t num_threads = num_rows;
- if (std::is_same<xpu, gpu>::value) {
- num_threads = num_rows * row_length;
- }
- Kernel<AdamDnsRspDnsKernel<req_type, xpu>, xpu>::Launch(s, num_threads,
- row_length, out_data, mean_data, var_data, weight_data, grad_idx, grad_val,
+ Kernel<AdamDnsRspDnsKernel<req_type>, xpu>::Launch(s, num_rows, row_length,
+ out_data, mean_data, var_data, weight_data, grad_idx, grad_val,
static_cast<DType>(param.clip_gradient), static_cast<DType>(param.beta1),
static_cast<DType>(param.beta2), static_cast<DType>(param.lr),
static_cast<DType>(param.wd), static_cast<DType>(param.epsilon),
@@ -892,8 +858,42 @@ inline void AdamUpdateRspRspRspImpl(const AdamParam& param,
var.data(), req, &out_blob);
}
-template<int req, typename xpu>
-struct AdamStdDnsRspDnsKernel;
+template<int req>
+struct AdamStdDnsRspDnsKernel {
+ template<typename DType, typename IType, typename RType>
+ MSHADOW_XINLINE static void Map(int i, const nnvm::dim_t row_length, DType* out_data,
+ DType* mean_data, DType* var_data, const DType* weight_data, const IType* grad_idx,
+ const DType* grad_data, const RType* prefix_sum, const DType clip_gradient,
+ const DType beta1, const DType beta2, const DType lr, const DType wd,
+ const DType epsilon, const DType rescale_grad) {
+ using namespace mshadow_op;
+ const bool non_zero = (i == 0) ? prefix_sum[0] > 0
+ : prefix_sum[i] > prefix_sum[i-1];
+
+ const index_t row_i = i * row_length;
+ const RType grad_i = (prefix_sum[i]-1) * row_length;
+ for (index_t j = 0; j < row_length; j++) {
+ const index_t data_i = row_i + j;
+ const DType grad_rescaled = non_zero ? static_cast<DType>(
+ grad_data[grad_i + j] * rescale_grad +
+ weight_data[data_i] * wd)
+ : static_cast<DType>(weight_data[data_i] * wd);
+ if (clip_gradient >= 0.0f) {
+ mean_data[data_i] = beta1 * mean_data[data_i] + (1.f - beta1) *
+ clip::Map(grad_rescaled, clip_gradient);
+ var_data[data_i] = beta2 * var_data[data_i] + (1.f - beta2) * square::Map(
+ clip::Map(grad_rescaled, clip_gradient));
+ } else {
+ mean_data[data_i] = beta1 * mean_data[data_i] + (1.f - beta1) * grad_rescaled;
+ var_data[data_i] = beta2 * var_data[data_i] +
+ (1.f - beta2) * square::Map(grad_rescaled);
+ }
+ KERNEL_ASSIGN(out_data[data_i], req, weight_data[data_i] - lr * mean_data[data_i] /
+ (square_root::Map(var_data[data_i]) + epsilon));
+ }
+ }
+};
+
template<typename xpu>
void AdamStdUpdateDnsRspDnsImpl(const AdamParam& param,
diff --git a/src/operator/optimizer_op.cc b/src/operator/optimizer_op.cc
index f7ccbbb739d..741092ad784 100644
--- a/src/operator/optimizer_op.cc
+++ b/src/operator/optimizer_op.cc
@@ -149,43 +149,6 @@ void SGDMomStdUpdateDnsRspDnsImpl<cpu>(const SGDMomParam& param,
});
}
-template<int req>
-struct AdamStdDnsRspDnsKernel<req, cpu> {
- template<typename DType, typename IType, typename RType>
- MSHADOW_XINLINE static void Map(int i, const nnvm::dim_t row_length, DType* out_data,
- DType* mean_data, DType* var_data, const DType* weight_data, const IType* grad_idx,
- const DType* grad_data, const RType* prefix_sum, const DType clip_gradient,
- const DType beta1, const DType beta2, const DType lr, const DType wd,
- const DType epsilon, const DType rescale_grad) {
- using namespace mshadow_op;
- const bool non_zero = (i == 0) ? prefix_sum[0] > 0
- : prefix_sum[i] > prefix_sum[i-1];
-
- const index_t row_i = i * row_length;
- const RType grad_i = (prefix_sum[i]-1) * row_length;
- for (index_t j = 0; j < row_length; j++) {
- const index_t data_i = row_i + j;
- const DType grad_rescaled = non_zero ? static_cast<DType>(
- grad_data[grad_i + j] * rescale_grad +
- weight_data[data_i] * wd)
- : static_cast<DType>(weight_data[data_i] * wd);
- if (clip_gradient >= 0.0f) {
- mean_data[data_i] = beta1 * mean_data[data_i] + (1.f - beta1) *
- clip::Map(grad_rescaled, clip_gradient);
- var_data[data_i] = beta2 * var_data[data_i] + (1.f - beta2) * square::Map(
- clip::Map(grad_rescaled, clip_gradient));
- } else {
- mean_data[data_i] = beta1 * mean_data[data_i] + (1.f - beta1) * grad_rescaled;
- var_data[data_i] = beta2 * var_data[data_i] +
- (1.f - beta2) * square::Map(grad_rescaled);
- }
- KERNEL_ASSIGN(out_data[data_i], req, weight_data[data_i] - lr * mean_data[data_i] /
- (square_root::Map(var_data[data_i]) + epsilon));
- }
- }
-};
-
-
template<>
void AdamStdUpdateDnsRspDnsImpl<cpu>(const AdamParam& param,
const OpContext& ctx,
@@ -231,7 +194,7 @@ void AdamStdUpdateDnsRspDnsImpl<cpu>(const AdamParam& param,
}
}
- Kernel<AdamStdDnsRspDnsKernel<req_type, cpu>, cpu>::Launch(s, num_rows, row_length,
+ Kernel<AdamStdDnsRspDnsKernel<req_type>, cpu>::Launch(s, num_rows, row_length,
out_data, mean_data, var_data, weight_data, grad_idx, grad_val, prefix_sum,
static_cast<DType>(param.clip_gradient), static_cast<DType>(param.beta1),
static_cast<DType>(param.beta2), static_cast<DType>(param.lr),
diff --git a/src/operator/optimizer_op.cu b/src/operator/optimizer_op.cu
index 18ee66a729c..c49af68a5f6 100644
--- a/src/operator/optimizer_op.cu
+++ b/src/operator/optimizer_op.cu
@@ -94,35 +94,6 @@ void SGDMomStdUpdateDnsRspDnsImpl<gpu>(const SGDMomParam& param,
});
}
-template<int req>
-struct AdamStdDnsRspDnsKernel<req, gpu> {
- template<typename DType, typename IType, typename RType>
- MSHADOW_XINLINE static void Map(int i, const nnvm::dim_t row_length, DType* out_data,
- DType* mean_data, DType* var_data, const DType* weight_data, const IType* grad_idx,
- const DType* grad_data, const RType* prefix_sum, const DType clip_gradient,
- const DType beta1, const DType beta2, const DType lr, const DType wd,
- const DType epsilon, const DType rescale_grad) {
- using namespace mshadow_op;
- using nnvm::dim_t;
- const dim_t row_id = i / row_length;
- const dim_t col_id = i % row_length;
- const bool non_zero = (row_id == 0) ? prefix_sum[0] > 0
- : prefix_sum[row_id] > prefix_sum[row_id - 1];
- const RType grad_offset = (prefix_sum[row_id] - 1) * row_length + col_id;
- DType grad_rescaled = non_zero ? static_cast<DType>(grad_data[grad_offset] * rescale_grad
- + weight_data[i] * wd)
- : static_cast<DType>(weight_data[i] * wd);
- if (clip_gradient >= 0.0f) {
- grad_rescaled = clip::Map(grad_rescaled, clip_gradient);
- }
- mean_data[i] = beta1 * mean_data[i] + (1.f - beta1) * grad_rescaled;
- var_data[i] = beta2 * var_data[i] +
- (1.f - beta2) * square::Map(grad_rescaled);
- KERNEL_ASSIGN(out_data[i], req, weight_data[i] - lr * mean_data[i] /
- (square_root::Map(var_data[i]) + epsilon));
- }
-};
-
template<>
void AdamStdUpdateDnsRspDnsImpl<gpu>(const AdamParam& param,
const OpContext& ctx,
@@ -151,8 +122,8 @@ void AdamStdUpdateDnsRspDnsImpl<gpu>(const AdamParam& param,
DType* mean_data = mean.dptr<DType>();
DType* var_data = var.dptr<DType>();
DType* out_data = out->dptr<DType>();
- const nnvm::dim_t num_rows = weight.shape_[0];
- const nnvm::dim_t row_length = weight.shape_.ProdShape(1, weight.ndim());
+ nnvm::dim_t num_rows = weight.shape_[0];
+ nnvm::dim_t row_length = weight.shape_.ProdShape(1, weight.ndim());
nnvm::dim_t* prefix_sum = NULL;
void* d_temp_storage = NULL;
size_t temp_storage_bytes = 0;
@@ -181,8 +152,8 @@ void AdamStdUpdateDnsRspDnsImpl<gpu>(const AdamParam& param,
Stream<gpu>::GetStream(s));
}
- Kernel<AdamStdDnsRspDnsKernel<req_type, gpu>, gpu>::Launch(s, weight.shape_.Size(),
- row_length, out_data, mean_data, var_data, weight_data, grad_idx, grad_val, prefix_sum,
+ Kernel<AdamStdDnsRspDnsKernel<req_type>, gpu>::Launch(s, num_rows, row_length,
+ out_data, mean_data, var_data, weight_data, grad_idx, grad_val, prefix_sum,
static_cast<DType>(param.clip_gradient), static_cast<DType>(param.beta1),
static_cast<DType>(param.beta2), static_cast<DType>(param.lr),
static_cast<DType>(param.wd), static_cast<DType>(param.epsilon),
diff --git a/src/profiler/profiler.h b/src/profiler/profiler.h
index b8d0e8ef340..768a0bc7d71 100644
--- a/src/profiler/profiler.h
+++ b/src/profiler/profiler.h
@@ -391,14 +391,6 @@ class Profiler {
return aggregate_stats_.get() != nullptr;
}
- /*!
- * \brief Whether aggregate stats are currently being recorded
- * \return true if aggregate stats are currently being recorded
- */
- inline bool AggregateRunning() const {
- return GetState() == kRunning && AggregateEnabled();
- }
-
public:
/*!
* \brief Constructor
diff --git a/tests/python-pytest/onnx/backend.py b/tests/python-pytest/onnx/backend.py
index 0e0a6a680b7..3b99563bccf 100644
--- a/tests/python-pytest/onnx/backend.py
+++ b/tests/python-pytest/onnx/backend.py
@@ -94,15 +94,12 @@ def run_node(cls, node, inputs, device='CPU'):
result obtained after running the operator
"""
graph = GraphProto()
- sym, arg_params, aux_params = graph.from_onnx(MXNetBackend.make_graph(node, inputs))
- data_names = [graph_input for graph_input in sym.list_inputs()
- if graph_input not in arg_params and graph_input not in aux_params]
+ sym, _ = graph.from_onnx(MXNetBackend.make_graph(node, inputs))
+ data_names = [i for i in sym.get_internals().list_inputs()]
data_shapes = []
dim_change_op_types = set(['ReduceMin', 'ReduceMax', 'ReduceMean',
'ReduceProd', 'ReduceSum', 'Slice', 'Pad',
- 'Squeeze', 'Upsample', 'Reshape', 'Conv',
- 'Concat', 'Softmax', 'Flatten', 'Transpose',
- 'GlobalAveragePool', 'GlobalMaxPool'])
+ 'Squeeze', 'Upsample', 'Reshape', 'Conv'])
# Adding extra dimension of batch_size 1 if the batch_size is different for multiple inputs.
for idx, input_name in enumerate(data_names):
@@ -126,10 +123,7 @@ def run_node(cls, node, inputs, device='CPU'):
mod.bind(for_training=False, data_shapes=data_shapes, label_shapes=None)
# initializing parameters for calculating result of each individual node
- if arg_params is None and aux_params is None:
- mod.init_params()
- else:
- mod.set_params(arg_params=arg_params, aux_params=aux_params)
+ mod.init_params()
data_forward = []
for idx, input_name in enumerate(data_names):
@@ -168,8 +162,8 @@ def prepare(cls, model, device='CPU', **kwargs):
used to run inference on the input model and return the result for comparison.
"""
graph = GraphProto()
- sym, arg_params, aux_params = graph.from_onnx(model.graph)
- return MXNetBackendRep(sym, arg_params, aux_params, device)
+ sym, params = graph.from_onnx(model.graph)
+ return MXNetBackendRep(sym, params, device)
@classmethod
def supports_device(cls, device):
diff --git a/tests/python-pytest/onnx/backend_rep.py b/tests/python-pytest/onnx/backend_rep.py
index 47ea6c1585a..a125086bce2 100644
--- a/tests/python-pytest/onnx/backend_rep.py
+++ b/tests/python-pytest/onnx/backend_rep.py
@@ -37,10 +37,9 @@
class MXNetBackendRep(BackendRep):
"""Running model inference on mxnet engine and return the result
to onnx test infrastructure for comparison."""
- def __init__(self, symbol, arg_params, aux_params, device):
+ def __init__(self, symbol, params, device):
self.symbol = symbol
- self.arg_params = arg_params
- self.aux_params = aux_params
+ self.params = params
self.device = device
def run(self, inputs, **kwargs):
@@ -68,7 +67,7 @@ def run(self, inputs, **kwargs):
label_names=None)
mod.bind(for_training=False, data_shapes=[('input_0', input_data.shape)],
label_shapes=None)
- mod.set_params(arg_params=self.arg_params, aux_params=self.aux_params)
+ mod.set_params(arg_params=self.params, aux_params=None)
# run inference
batch = namedtuple('Batch', ['data'])
diff --git a/tests/python-pytest/onnx/onnx_backend_test.py b/tests/python-pytest/onnx/onnx_backend_test.py
index 4ea31e5aac9..28e2aaefcdd 100644
--- a/tests/python-pytest/onnx/onnx_backend_test.py
+++ b/tests/python-pytest/onnx/onnx_backend_test.py
@@ -34,7 +34,7 @@
BACKEND_TEST = onnx.backend.test.BackendTest(mxnet_backend, __name__)
-IMPLEMENTED_OPERATORS_TEST = [
+IMPLEMENTED_OPERATORS = [
#Generator Functions
#'test_constant*', # Identity Function
#'test_random_uniform',
@@ -57,40 +57,37 @@
'test_floor',
## Joining and spliting
- 'test_concat',
+ #'test_concat.*', #---Failing test
#Basic neural network functions
'test_sigmoid',
'test_relu',
- 'test_constant_pad',
- 'test_edge_pad',
- 'test_reflect_pad',
+ #'test_constant_pad',
+ #'test_edge_pad',
+ #'test_reflect_pad',
'test_matmul',
'test_leakyrelu',
'test_elu',
- 'test_softmax_example',
- 'test_softmax_large_number',
- 'test_softmax_axis_2',
+ #'test_softmax*',
'test_conv',
'test_basic_conv',
- 'test_transpose',
- 'test_globalmaxpool',
- 'test_globalaveragepool',
- #'test_batch_norm', - tests to be added
- #'test_gather',
+ #'test_globalmaxpool',
+ #'test_globalaveragepool',
+ #'test_batch_norm',
#Changing shape and type.
'test_reshape_',
- 'test_cast',
+ #'test_AvgPool2D*',
+ #'test_MaxPool2D*',
+ #'test_cast',
#'test_split',
'test_slice_cpu',
'test_default_axes', #make PR against onnx to fix the test name(grep-able)
'test_slice_neg',
#'test_slice_start_out_of_bounds',
#'test_slice_end_out_of_bounds',
- #'test_transpose',
+ #'test_transpose*',
'test_squeeze_',
- 'test_flatten_default',
#Powers
'test_reciprocal',
@@ -106,62 +103,12 @@
'test_argmax',
'test_argmin',
'test_max',
- 'test_min',
-
- #pytorch operator tests
- #'test_operator_chunk',
- #'test_operator_clip',
- 'test_operator_conv',
- #'test_operator_equal',
- 'test_operator_exp',
- #'test_operator_flatten',
- #'test_operator_max',
- 'test_operator_maxpool',
- 'test_operator_non_float_params',
- 'test_operator_params',
- 'test_operator_permute2',
- #'test_operator_transpose',
- #'test_operator_view'
+ 'test_min'
]
-BASIC_MODEL_TESTS = [
- 'test_AvgPool2D',
- 'test_BatchNorm',
- 'test_ConstantPad2d'
- 'test_Conv2d',
- 'test_ELU',
- 'test_LeakyReLU',
- 'test_MaxPool',
- 'test_PReLU',
- 'test_ReLU',
- 'test_Sigmoid',
- 'test_Softmax',
- 'test_softmax_functional',
- 'test_softmax_lastdim',
- 'test_Tanh'
- ]
-
-STANDARD_MODEL = [
- 'test_bvlc_alexnet',
- 'test_densenet121',
- #'test_inception_v1',
- #'test_inception_v2',
- 'test_resnet50',
- #'test_shufflenet',
- 'test_squeezenet',
- 'test_vgg16',
- 'test_vgg19'
- ]
-
-for op_test in IMPLEMENTED_OPERATORS_TEST:
+for op_test in IMPLEMENTED_OPERATORS:
BACKEND_TEST.include(op_test)
-for std_model_test in STANDARD_MODEL:
- BACKEND_TEST.include(std_model_test)
-
-for basic_model_test in BASIC_MODEL_TESTS:
- BACKEND_TEST.include(basic_model_test)
-
# import all test cases at global scope to make them visible to python.unittest
globals().update(BACKEND_TEST.enable_report().test_cases)
diff --git a/tests/python-pytest/onnx/onnx_test.py b/tests/python-pytest/onnx/onnx_test.py
index ddc633e28f6..016490a4c4b 100644
--- a/tests/python-pytest/onnx/onnx_test.py
+++ b/tests/python-pytest/onnx/onnx_test.py
@@ -21,37 +21,19 @@
ONNX backend test framework. Once we have PRs on the ONNX repo and get
those PRs merged, this file will get EOL'ed.
"""
-# pylint: disable=too-many-locals,wrong-import-position,import-error
from __future__ import absolute_import
import sys
import os
import unittest
import logging
import hashlib
-import tarfile
-from collections import namedtuple
import numpy as np
import numpy.testing as npt
from onnx import helper
-from onnx import numpy_helper
-from onnx import TensorProto
-from mxnet.test_utils import download
-from mxnet.contrib import onnx as onnx_mxnet
-import mxnet as mx
+import backend as mxnet_backend
CURR_PATH = os.path.dirname(os.path.abspath(os.path.expanduser(__file__)))
sys.path.insert(0, os.path.join(CURR_PATH, '../../python/unittest'))
from common import with_seed
-import backend as mxnet_backend
-
-
-URLS = {
- 'bvlc_googlenet' :
- 'https://s3.amazonaws.com/onnx-mxnet/model-zoo/bvlc_googlenet.tar.gz',
- 'bvlc_reference_caffenet' :
- 'https://s3.amazonaws.com/onnx-mxnet/model-zoo/bvlc_reference_caffenet.tar.gz',
- 'bvlc_reference_rcnn_ilsvrc13' :
- 'https://s3.amazonaws.com/onnx-mxnet/model-zoo/bvlc_reference_rcnn_ilsvrc13.tar.gz',
-}
@with_seed()
def test_reduce_max():
@@ -111,9 +93,9 @@ def test_super_resolution_example():
sys.path.insert(0, os.path.join(CURR_PATH, '../../../example/onnx/'))
import super_resolution
- sym, arg_params, aux_params = super_resolution.import_onnx()
+ sym, params = super_resolution.import_onnx()
assert sym is not None
- assert arg_params is not None
+ assert params is not None
inputs = sym.list_inputs()
assert len(inputs) == 9
@@ -134,7 +116,7 @@ def test_super_resolution_example():
'transpose0']):
assert key_item in attrs_keys
- param_keys = arg_params.keys()
+ param_keys = params.keys()
assert len(param_keys) == 8
for i, param_item in enumerate(['param_5', 'param_4', 'param_7', 'param_6',
'param_1', 'param_0', 'param_3', 'param_2']):
@@ -144,111 +126,11 @@ def test_super_resolution_example():
output_img_dim = 672
input_image, img_cb, img_cr = super_resolution.get_test_image()
- result_img = super_resolution.perform_inference(sym, arg_params, aux_params,
- input_image, img_cb, img_cr)
+ result_img = super_resolution.perform_inference(sym, params, input_image,
+ img_cb, img_cr)
assert hashlib.md5(result_img.tobytes()).hexdigest() == '0d98393a49b1d9942106a2ed89d1e854'
assert result_img.size == (output_img_dim, output_img_dim)
-def get_test_files(name):
- """Extract tar file and returns model path and input, output data"""
- tar_name = download(URLS.get(name), dirname=CURR_PATH.__str__())
- # extract tar file
- tar_path = os.path.join(CURR_PATH, tar_name)
- tar = tarfile.open(tar_path.__str__(), "r:*")
- tar.extractall(path=CURR_PATH.__str__())
- tar.close()
- data_dir = os.path.join(CURR_PATH, name)
- model_path = os.path.join(data_dir, 'model.onnx')
-
- inputs = []
- outputs = []
- # get test files
- for test_file in os.listdir(data_dir):
- case_dir = os.path.join(data_dir, test_file)
- # skip the non-dir files
- if not os.path.isdir(case_dir):
- continue
- input_file = os.path.join(case_dir, 'input_0.pb')
- input_tensor = TensorProto()
- with open(input_file, 'rb') as proto_file:
- input_tensor.ParseFromString(proto_file.read())
- inputs.append(numpy_helper.to_array(input_tensor))
-
- output_tensor = TensorProto()
- output_file = os.path.join(case_dir, 'output_0.pb')
- with open(output_file, 'rb') as proto_file:
- output_tensor.ParseFromString(proto_file.read())
- outputs.append(numpy_helper.to_array(output_tensor))
-
- return model_path, inputs, outputs
-
-def test_bvlc_googlenet():
- """ Tests Googlenet model"""
- model_path, inputs, outputs = get_test_files('bvlc_googlenet')
- logging.info("Translating Googlenet model from ONNX to Mxnet")
- sym, arg_params, aux_params = onnx_mxnet.import_model(model_path)
-
- # run test for each test file
- for input_data, output_data in zip(inputs, outputs):
- # create module
- mod = mx.mod.Module(symbol=sym, data_names=['input_0'], context=mx.cpu(), label_names=None)
- mod.bind(for_training=False, data_shapes=[('input_0', input_data.shape)], label_shapes=None)
- mod.set_params(arg_params=arg_params, aux_params=aux_params,
- allow_missing=True, allow_extra=True)
- # run inference
- batch = namedtuple('Batch', ['data'])
- mod.forward(batch([mx.nd.array(input_data)]), is_train=False)
-
- # verify the results
- npt.assert_equal(mod.get_outputs()[0].shape, output_data.shape)
- npt.assert_almost_equal(output_data, mod.get_outputs()[0].asnumpy(), decimal=3)
- logging.info("Googlenet model conversion Successful")
-
-def test_bvlc_reference_caffenet():
- """Tests the bvlc cafenet model"""
- model_path, inputs, outputs = get_test_files('bvlc_reference_caffenet')
- logging.info("Translating Caffenet model from ONNX to Mxnet")
- sym, arg_params, aux_params = onnx_mxnet.import_model(model_path)
-
- # run test for each test file
- for input_data, output_data in zip(inputs, outputs):
- # create module
- mod = mx.mod.Module(symbol=sym, data_names=['input_0'], context=mx.cpu(), label_names=None)
- mod.bind(for_training=False, data_shapes=[('input_0', input_data.shape)], label_shapes=None)
- mod.set_params(arg_params=arg_params, aux_params=aux_params,
- allow_missing=True, allow_extra=True)
- # run inference
- batch = namedtuple('Batch', ['data'])
- mod.forward(batch([mx.nd.array(input_data)]), is_train=False)
-
- # verify the results
- npt.assert_equal(mod.get_outputs()[0].shape, output_data.shape)
- npt.assert_almost_equal(output_data, mod.get_outputs()[0].asnumpy(), decimal=3)
- logging.info("Caffenet model conversion Successful")
-
-def test_bvlc_rcnn_ilsvrc13():
- """Tests the bvlc rcnn model"""
- model_path, inputs, outputs = get_test_files('bvlc_reference_rcnn_ilsvrc13')
- logging.info("Translating rcnn_ilsvrc13 model from ONNX to Mxnet")
- sym, arg_params, aux_params = onnx_mxnet.import_model(model_path)
-
- # run test for each test file
- for input_data, output_data in zip(inputs, outputs):
- # create module
- mod = mx.mod.Module(symbol=sym, data_names=['input_0'], context=mx.cpu(), label_names=None)
- mod.bind(for_training=False, data_shapes=[('input_0', input_data.shape)], label_shapes=None)
- mod.set_params(arg_params=arg_params, aux_params=aux_params,
- allow_missing=True, allow_extra=True)
- # run inference
- batch = namedtuple('Batch', ['data'])
- mod.forward(batch([mx.nd.array(input_data)]), is_train=False)
-
- # verify the results
- npt.assert_equal(mod.get_outputs()[0].shape, output_data.shape)
- npt.assert_almost_equal(output_data, mod.get_outputs()[0].asnumpy(), decimal=3)
- logging.info("rcnn_ilsvrc13 model conversion Successful")
-
-
if __name__ == '__main__':
unittest.main()
diff --git a/tests/python/unittest/test_gluon.py b/tests/python/unittest/test_gluon.py
index 952fdf7e366..ba2e7aba9f6 100644
--- a/tests/python/unittest/test_gluon.py
+++ b/tests/python/unittest/test_gluon.py
@@ -82,16 +82,16 @@ def hybrid_forward(self, F, x, const):
@with_seed()
def test_parameter_sharing():
class Net(gluon.Block):
- def __init__(self, in_units=0, **kwargs):
+ def __init__(self, **kwargs):
super(Net, self).__init__(**kwargs)
with self.name_scope():
- self.dense0 = nn.Dense(5, in_units=in_units)
- self.dense1 = nn.Dense(5, in_units=in_units)
+ self.dense0 = nn.Dense(5, in_units=5)
+ self.dense1 = nn.Dense(5, in_units=5)
def forward(self, x):
return self.dense1(self.dense0(x))
- net1 = Net(prefix='net1_', in_units=5)
+ net1 = Net(prefix='net1_')
net2 = Net(prefix='net2_', params=net1.collect_params())
net1.collect_params().initialize()
net2(mx.nd.zeros((3, 5)))
@@ -101,16 +101,6 @@ def forward(self, x):
net3 = Net(prefix='net3_')
net3.load_params('net1.params', mx.cpu())
- net4 = Net(prefix='net4_')
- net5 = Net(prefix='net5_', in_units=5, params=net4.collect_params())
- net4.collect_params().initialize()
- net5(mx.nd.zeros((3, 5)))
-
- net4.save_params('net4.params')
-
- net6 = Net(prefix='net6_')
- net6.load_params('net4.params', mx.cpu())
-
@with_seed()
def test_parameter_str():
diff --git a/tests/python/unittest/test_io.py b/tests/python/unittest/test_io.py
index e986ae7bf71..4e23a22e09e 100644
--- a/tests/python/unittest/test_io.py
+++ b/tests/python/unittest/test_io.py
@@ -219,9 +219,7 @@ def check_libSVMIter_synthetic():
i = 0
for batch in iter(data_train):
expected = first.asnumpy() if i == 0 else second.asnumpy()
- data = data_train.getdata()
- data.check_format(True)
- assert_almost_equal(data.asnumpy(), expected)
+ assert_almost_equal(data_train.getdata().asnumpy(), expected)
i += 1
def check_libSVMIter_news_data():
@@ -229,7 +227,7 @@ def check_libSVMIter_news_data():
'name': 'news20.t',
'origin_name': 'news20.t.bz2',
'url': "https://apache-mxnet.s3-accelerate.dualstack.amazonaws.com/gluon/dataset/news20.t.bz2",
- 'feature_dim': 62060 + 1,
+ 'feature_dim': 62060,
'num_classes': 20,
'num_examples': 3993,
}
@@ -245,11 +243,8 @@ def check_libSVMIter_news_data():
num_batches = 0
for batch in data_train:
# check the range of labels
- data = batch.data[0]
- label = batch.label[0]
- data.check_format(True)
- assert(np.sum(label.asnumpy() > 20) == 0)
- assert(np.sum(label.asnumpy() <= 0) == 0)
+ assert(np.sum(batch.label[0].asnumpy() > 20) == 0)
+ assert(np.sum(batch.label[0].asnumpy() <= 0) == 0)
num_batches += 1
expected_num_batches = num_examples / batch_size
assert(num_batches == int(expected_num_batches)), num_batches
diff --git a/tests/python/unittest/test_operator.py b/tests/python/unittest/test_operator.py
index 561e6952557..2486be04a52 100644
--- a/tests/python/unittest/test_operator.py
+++ b/tests/python/unittest/test_operator.py
@@ -24,7 +24,6 @@
import itertools
from numpy.testing import assert_allclose, assert_array_equal
from mxnet.test_utils import *
-from mxnet.base import py_str
from common import setup_module, with_seed
import unittest
@@ -490,9 +489,7 @@ def frelu_grad(x):
check_symbolic_backward(y, [xa], [np.ones(shape)], [ga])
-# NOTE(haojin2): Skipping the numeric check tests for float16 data type due to precision issues,
-# the analytical checks are still performed on each and every data type to verify the correctness.
-@with_seed()
+@with_seed(1234)
def test_leaky_relu():
def fleaky_relu(x, act_type, slope=0.25):
neg_indices = x < 0
@@ -512,27 +509,22 @@ def fleaky_relu_grad(grad, x, y, act_type, slope=0.25):
return out * grad
shape = (3, 4)
x = mx.symbol.Variable("x")
- slp = 0.25
+ slp = 0.0625
for dtype in [np.float16, np.float32, np.float64]:
- xa = np.random.uniform(low=-1.0,high=1.0,size=shape).astype(dtype)
+ xa = np.random.uniform(low=-1.0,high=-0.2,size=shape).astype(dtype)
eps = 1e-4
- rtol = 1e-4
- atol = 1e-3
xa[abs(xa) < eps] = 1.0
- for act_type in ['elu', 'leaky']:
+ # eps = 1e-2 if dtype is np.float16 else 1e-4
+ for act_type in ['leaky']:
y = mx.symbol.LeakyReLU(data=x, slope=slp, act_type=act_type)
ya = fleaky_relu(xa, slope=slp, act_type=act_type)
ga = fleaky_relu_grad(np.ones(shape), xa, ya, slope=slp, act_type=act_type)
- # Skip numeric check for float16 type to get rid of flaky behavior
- if dtype is not np.float16:
- check_numeric_gradient(y, [xa], numeric_eps=eps, rtol=rtol, atol=atol, dtype=dtype)
- check_symbolic_forward(y, [xa], [ya], rtol=rtol, atol=atol, dtype=dtype)
- check_symbolic_backward(y, [xa], [np.ones(shape)], [ga], rtol=rtol, atol=atol, dtype=dtype)
+ check_numeric_gradient(y, [xa], numeric_eps=eps, rtol=1e-4, atol=1e-4)
+ check_symbolic_forward(y, [xa], [ya], rtol=eps, atol=1e-5, dtype=dtype)
+ check_symbolic_backward(y, [xa], [np.ones(shape)], [ga], rtol=eps, atol=1e-5, dtype=dtype)
-# NOTE(haojin2): Skipping the numeric check tests for float16 data type due to precision issues,
-# the analytical checks are still performed on each and every data type to verify the correctness.
-@with_seed()
+@with_seed(1234)
def test_prelu():
def fprelu(x, gamma):
pos_indices = x > 0
@@ -556,20 +548,17 @@ def fprelu_grad(x, y, gamma):
x = mx.symbol.Variable("x")
gamma = mx.symbol.Variable("gamma")
for dtype in [np.float16, np.float32, np.float64]:
- for gam in [np.array([0.1, 0.2, 0.3, 0.4], dtype=dtype)]:
+ for gam in [np.array([0.1], dtype=dtype), np.array([0.1, 0.2, 0.3, 0.4], dtype=dtype)]:
xa = np.random.uniform(low=-1.0,high=1.0,size=shape).astype(dtype)
- rtol = 1e-3
- atol = 1e-3
eps = 1e-4
xa[abs(xa) < eps] = 1.0
y = mx.symbol.LeakyReLU(data=x, gamma=gamma, act_type='prelu')
ya = fprelu(xa, gam)
g_xa, g_gam = fprelu_grad(xa, ya, gamma=gam)
- # Skip numeric check for float16 type to get rid of flaky behavior
- if dtype is not np.float16:
- check_numeric_gradient(y, [xa, gam], numeric_eps=eps, rtol=rtol, atol=atol, dtype=dtype)
- check_symbolic_forward(y, [xa, gam], [ya], rtol=rtol, atol=atol, dtype=dtype)
- check_symbolic_backward(y, [xa, gam], [np.ones(shape), np.ones(gam.shape)], [g_xa, g_gam], rtol=rtol, atol=atol, dtype=dtype)
+ check_numeric_gradient(y, [xa, gam], numeric_eps=eps, rtol=1e-3, atol=1e-4)
+ check_symbolic_forward(y, [xa, gam], [ya], rtol=1e-3, atol=1e-20)
+ check_symbolic_backward(y, [xa, gam], [np.ones(shape)], [g_xa], rtol=1e-3, atol=1e-20)
+
@with_seed()
def test_sigmoid():
@@ -5410,30 +5399,6 @@ def f(x, a, b, c):
check_numeric_gradient(quad_sym, [data_np], atol=0.001)
-def test_op_output_names_monitor():
- def check_name(op_sym, expected_names):
- output_names = []
-
- def get_output_names_callback(name, arr):
- output_names.append(py_str(name))
-
- op_exe = op_sym.simple_bind(ctx=mx.current_context(), grad_req='null')
- op_exe.set_monitor_callback(get_output_names_callback)
- op_exe.forward()
- for output_name, expected_name in zip(output_names, expected_names):
- assert output_name == expected_name
-
- data = mx.sym.Variable('data', shape=(10, 3, 10, 10))
- conv_sym = mx.sym.Convolution(data, kernel=(2, 2), num_filter=1, name='conv')
- check_name(conv_sym, ['conv_output'])
-
- fc_sym = mx.sym.FullyConnected(data, num_hidden=10, name='fc')
- check_name(fc_sym, ['fc_output'])
-
- lrn_sym = mx.sym.LRN(data, nsize=1, name='lrn')
- check_name(lrn_sym, ['lrn_output', 'lrn_tmp_norm'])
-
-
if __name__ == '__main__':
import nose
nose.runmodule()
diff --git a/tests/python/unittest/test_optimizer.py b/tests/python/unittest/test_optimizer.py
index bbd7845f66f..f71e2c81e27 100644
--- a/tests/python/unittest/test_optimizer.py
+++ b/tests/python/unittest/test_optimizer.py
@@ -543,13 +543,13 @@ def test_ftml():
class PyAdam(mx.optimizer.Optimizer):
"""python reference implemenation of adam"""
def __init__(self, learning_rate=0.001, beta1=0.9, beta2=0.999, epsilon=1e-8,
- decay_factor=(1 - 1e-8), lazy_update=False, **kwargs):
+ decay_factor=(1 - 1e-8), sparse_update=False, **kwargs):
super(PyAdam, self).__init__(learning_rate=learning_rate, **kwargs)
self.beta1 = beta1
self.beta2 = beta2
self.epsilon = epsilon
self.decay_factor = decay_factor
- self.lazy_update = lazy_update
+ self.sparse_update = sparse_update
def create_state(self, index, weight):
"""Create additional optimizer state: mean, variance
@@ -595,7 +595,7 @@ def update(self, index, weight, grad, state):
# check row slices of all zeros
all_zeros = mx.test_utils.almost_equal(grad[row].asnumpy(), np.zeros_like(grad[row].asnumpy()))
# skip zeros during sparse update
- if all_zeros and self.lazy_update:
+ if all_zeros and self.sparse_update:
continue
grad[row] = grad[row] * self.rescale_grad + wd * weight[row]
# clip gradients
@@ -638,7 +638,7 @@ def test_adam():
compare_optimizer(opt1(**kwarg), opt2(**kwarg), shape, dtype,
rtol=1e-4, atol=2e-5)
# atol 2e-5 needed to pass with seed 781809840
- compare_optimizer(opt1(lazy_update=True, **kwarg), opt2(**kwarg), shape,
+ compare_optimizer(opt1(sparse_update=True, **kwarg), opt2(**kwarg), shape,
dtype, w_stype='row_sparse', g_stype='row_sparse',
rtol=1e-4, atol=2e-5)
compare_optimizer(opt1(**kwarg), opt2(lazy_update=False, **kwarg), shape,
@@ -883,12 +883,12 @@ class PyFtrl(mx.optimizer.Optimizer):
\\eta_{t,i} = \\frac{learningrate}{\\beta+\\sqrt{\\sum_{s=1}^tg_{s,i}^t}}
"""
- def __init__(self, lamda1=0.01, learning_rate=0.1, beta=1, lazy_update=False, **kwargs):
+ def __init__(self, lamda1=0.01, learning_rate=0.1, beta=1, sparse_update=False, **kwargs):
super(PyFtrl, self).__init__(**kwargs)
self.lamda1 = lamda1
self.beta = beta
self.lr = learning_rate
- self.lazy_update = lazy_update
+ self.sparse_update = sparse_update
def create_state(self, index, weight):
return (mx.nd.zeros(weight.shape, weight.context, dtype=weight.dtype), # dn
@@ -903,7 +903,7 @@ def update(self, index, weight, grad, state):
dn, n = state
for row in range(num_rows):
all_zeros = mx.test_utils.almost_equal(grad[row].asnumpy(), np.zeros_like(grad[row].asnumpy()))
- if all_zeros and self.lazy_update:
+ if all_zeros and self.sparse_update:
continue
grad[row] = grad[row] * self.rescale_grad
if self.clip_gradient is not None:
@@ -933,7 +933,7 @@ def test_ftrl():
{'clip_gradient': 0.5, 'wd': 0.07, 'lamda1': 1.0}]
for kwarg in kwargs:
compare_optimizer(opt1(**kwarg), opt2(**kwarg), shape, np.float32)
- compare_optimizer(opt1(lazy_update=True, **kwarg), opt2(**kwarg), shape,
+ compare_optimizer(opt1(sparse_update=True, **kwarg), opt2(**kwarg), shape,
np.float32, w_stype='row_sparse', g_stype='row_sparse')
@with_seed(1234)
diff --git a/tests/python/unittest/test_sparse_operator.py b/tests/python/unittest/test_sparse_operator.py
index 9417df31748..a8bf5a5ed3c 100644
--- a/tests/python/unittest/test_sparse_operator.py
+++ b/tests/python/unittest/test_sparse_operator.py
@@ -1820,21 +1820,6 @@ def check_scatter_ops(name, shape, lhs_stype, rhs_stype, forward_mxnet_call, for
lambda l, r: l + r,
rhs_is_scalar=True, verbose=False, density=0.5)
-@with_seed()
-def test_mkldnn_sparse():
- # This test is trying to create a race condition describedd in
- # https://github.com/apache/incubator-mxnet/issues/10189
- arr = mx.nd.random.uniform(shape=(10, 10, 32, 32))
- weight1 = mx.nd.random.uniform(shape=(10, 10, 3, 3))
- arr = mx.nd.Convolution(data=arr, weight=weight1, no_bias=True, kernel=(3, 3), num_filter=10)
-
- rs_arr = mx.nd.sparse.row_sparse_array((mx.nd.zeros_like(arr), np.arange(arr.shape[0])))
- weight2 = mx.nd.random.uniform(shape=(10, np.prod(arr.shape[1:4])))
- fc_res = mx.nd.FullyConnected(data=arr, weight=weight2, no_bias=True, num_hidden=10)
- sum_res = mx.nd.elemwise_sub(arr, rs_arr)
- res1 = np.dot(mx.nd.flatten(sum_res).asnumpy(), weight2.asnumpy().T)
- print(res1 - fc_res.asnumpy())
- almost_equal(res1, fc_res.asnumpy())
@with_seed()
def test_sparse_nd_where():
@@ -1933,26 +1918,6 @@ def test_where_numeric_gradient(shape):
test_where_helper((5, 9))
test_where_numeric_gradient((5, 9))
-@with_seed()
-def test_sparse_quadratic_function():
- def f(x, a, b, c):
- return a * x**2 + b * x + c
-
- def check_sparse_quadratic_function(a, b, c, expected_stype):
- # check forward and compare the result with dense op
- ndim = 2
- shape = rand_shape_nd(ndim, 5)
- data = rand_ndarray(shape=shape, stype='csr')
- data_np = data.asnumpy()
- expected = f(data_np, a, b, c)
- output = mx.nd.contrib.quadratic(data, a=a, b=b, c=c)
- assert(output.stype == expected_stype)
- assert_almost_equal(output.asnumpy(), expected)
-
- a = np.random.random_sample()
- b = np.random.random_sample()
- check_sparse_quadratic_function(a, b, 0.0, 'csr')
- check_sparse_quadratic_function(a, b, 1.0, 'default')
if __name__ == '__main__':
import nose
----------------------------------------------------------------
This is an automated message from the Apache Git Service.
To respond to the message, please log on GitHub and use the
URL above to go to the specific comment.
For queries about this service, please contact Infrastructure at:
users@infra.apache.org
With regards,
Apache Git Services