You are viewing a plain text version of this content. The canonical link for it is here.
Posted to reviews@spark.apache.org by KyleLi1985 <gi...@git.apache.org> on 2018/11/23 14:53:48 UTC
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
GitHub user KyleLi1985 opened a pull request:
https://github.com/apache/spark/pull/23126
[SPARK-26158] [MLLIB] fix covariance accuracy problem for DenseVector
## What changes were proposed in this pull request?
Enhance accuracy of the covariance logic in RowMatrix for function computeCovariance
## How was this patch tested?
Unit test
Accuracy test
You can merge this pull request into a Git repository by running:
$ git pull https://github.com/KyleLi1985/spark master
Alternatively you can review and apply these changes as the patch at:
https://github.com/apache/spark/pull/23126.patch
To close this pull request, make a commit to your master/trunk branch
with (at least) the following in the commit message:
This closes #23126
----
commit a8bfdfffbe82a77943adb4bf84ca939d786afc8a
Author: 李亮 <li...@...>
Date: 2018-11-23T14:35:27Z
fix covariance accuracy problem for DenseVector
----
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237532703
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,82 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+
+ val n = v.size
+ val na = Array.ofDim[Double](n)
+ val means = bc.value
+ if (v.isInstanceOf[DenseVector]) {
+ v.foreachActive{(index, value) =>
--- End diff --
This scalastyle not find my environment . We need consider sparse vector in here. If input some of sparse vector, we need to keep a right result. But, actually the sparse vector override the toArray function. So, update the newest commit.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
Compare Spark computeCovariance function in RowMatrix for DenseVector and Numpy's function cov,
Find two problem, below is the result:
1)The Spark function computeCovariance in RowMatrix is not accuracy
input data
1.0,2.0,3.0,4.0,5.0
2.0,3.0,1.0,2.0,6.0
Numpy function cov result:
[[2.5 1.75]
[ 1.75 3.7 ]]
RowMatrix function computeCovariance result:
2.5 1.75
1.75 3.700000000000001
2)For some input case, the result is not good
generate input data by below logic
data1 = np.random.normal(loc=100000, scale=0.000009, size=10000000)
data2 = np.random.normal(loc=200000, scale=0.000002,size=10000000)
Numpy function cov result:
[[ 8.10536442e-11 -4.35439574e-15]
[ -4.35439574e-15 3.99928264e-12]]
RowMatrix function computeCovariance result:
-0.0027484893798828125 0.001491546630859375
0.001491546630859375 8.087158203125E-4
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
Ok, I will do it later
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237130345
--- Diff: mllib/src/test/java/org/apache/spark/ml/feature/JavaPCASuite.java ---
@@ -67,7 +66,7 @@ public void testPCA() {
JavaRDD<Vector> dataRDD = jsc.parallelize(points, 2);
RowMatrix mat = new RowMatrix(dataRDD.map(
- (Vector vector) -> (org.apache.spark.mllib.linalg.Vector) new DenseVector(vector.toArray())
+ (Vector vector) -> org.apache.spark.mllib.linalg.Vectors.fromML(vector)
--- End diff --
I don't think that helps a caller much as they don't know what the implication of this is. I'd leave it for now.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by SparkQA <gi...@git.apache.org>.
Github user SparkQA commented on the issue:
https://github.com/apache/spark/pull/23126
**[Test build #4441 has finished](https://amplab.cs.berkeley.edu/jenkins/job/NewSparkPullRequestBuilder/4441/testReport)** for PR 23126 at commit [`6fca901`](https://github.com/apache/spark/commit/6fca901dfdad230fa7a4e1079a3f48be826276a3).
* This patch **fails Spark unit tests**.
* This patch merges cleanly.
* This patch adds no public classes.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by AmplabJenkins <gi...@git.apache.org>.
Github user AmplabJenkins commented on the issue:
https://github.com/apache/spark/pull/23126
Can one of the admins verify this patch?
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by SparkQA <gi...@git.apache.org>.
Github user SparkQA commented on the issue:
https://github.com/apache/spark/pull/23126
**[Test build #4441 has started](https://amplab.cs.berkeley.edu/jenkins/job/NewSparkPullRequestBuilder/4441/testReport)** for PR 23126 at commit [`6fca901`](https://github.com/apache/spark/commit/6fca901dfdad230fa7a4e1079a3f48be826276a3).
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
It would be better, update the commit
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on the issue:
https://github.com/apache/spark/pull/23126
Merged to master
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r236927721
--- Diff: mllib/src/test/java/org/apache/spark/ml/feature/JavaPCASuite.java ---
@@ -67,7 +66,7 @@ public void testPCA() {
JavaRDD<Vector> dataRDD = jsc.parallelize(points, 2);
RowMatrix mat = new RowMatrix(dataRDD.map(
- (Vector vector) -> (org.apache.spark.mllib.linalg.Vector) new DenseVector(vector.toArray())
+ (Vector vector) -> org.apache.spark.mllib.linalg.Vectors.fromML(vector)
--- End diff --
Sure, as you said, if the **first item** is sparse vector, it will align with computeSparseVectorCovariance logic, and if the **first item** is a dense vector, it will align with computeDenseVectorCovariance logic. The rest is user's choice.
But, maybe we can add some notes into the annotation of function computeCovariance,
give user some notes, like:
/**
* Computes the covariance matrix, treating each row as an observation.
*
* Note:
* When the first row is DenseVector, we use the computeDenseVectorCovariance
* to calculate the covariance matrix, and if the first row is SparseVector, we
* use the computeSparseVectorCovariance to calculate the covariance matrix
*
* @return a local dense matrix of size n x n
*
* @note This cannot be computed on matrices with more than 65535 columns.
*/
@Since("1.0.0")
def computeCovariance(): Matrix = {
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
After add this commit
We get the result for RowMatrix computeCovariance function:
For the input data
1.0,2.0,3.0,4.0,5.0
2.0,3.0,1.0,2.0,6.0
RowMatrix function computeCovariance result:
2.5 1.75
1.75 3.7
For the input data generated by
data1 = np.random.normal(loc=100000, scale=0.000009, size=10000000)
data2 = np.random.normal(loc=200000, scale=0.000002,size=10000000)
RowMatrix function computeCovariance result:
8.109505250896888E-11 -5.003160564607658E-15
-5.003160564607658E-15 4.08276584628234E-12
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by SparkQA <gi...@git.apache.org>.
Github user SparkQA commented on the issue:
https://github.com/apache/spark/pull/23126
**[Test build #4445 has started](https://amplab.cs.berkeley.edu/jenkins/job/NewSparkPullRequestBuilder/4445/testReport)** for PR 23126 at commit [`17117d7`](https://github.com/apache/spark/commit/17117d70ae0e26b5a99c6ea5a572cfbaf83080ab).
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237556042
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,82 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+
+ val n = v.size
+ val na = Array.ofDim[Double](n)
+ val means = bc.value
+ if (v.isInstanceOf[DenseVector]) {
+ v.foreachActive{(index, value) =>
--- End diff --
Yeah, because it hasn't subtracted the mean from one of the values in the Spark vector. that's the general issue with centering a sparse vector: it becomes dense!
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by AmplabJenkins <gi...@git.apache.org>.
Github user AmplabJenkins commented on the issue:
https://github.com/apache/spark/pull/23126
Can one of the admins verify this patch?
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by AmplabJenkins <gi...@git.apache.org>.
Github user AmplabJenkins commented on the issue:
https://github.com/apache/spark/pull/23126
Can one of the admins verify this patch?
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r236927771
--- Diff: mllib/src/test/scala/org/apache/spark/mllib/linalg/distributed/RowMatrixSuite.scala ---
@@ -266,6 +266,16 @@ class RowMatrixSuite extends SparkFunSuite with MLlibTestSparkContext {
}
}
+ test("dense vector covariance accuracy (SPARK-26158)") {
+ val rdd1 = sc.parallelize(Array(100000.000004, 100000.000012, 99999.9999931, 99999.9999977))
--- End diff --
handy thing
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r236658063
--- Diff: mllib/src/test/java/org/apache/spark/ml/feature/JavaPCASuite.java ---
@@ -67,7 +66,7 @@ public void testPCA() {
JavaRDD<Vector> dataRDD = jsc.parallelize(points, 2);
RowMatrix mat = new RowMatrix(dataRDD.map(
- (Vector vector) -> (org.apache.spark.mllib.linalg.Vector) new DenseVector(vector.toArray())
+ (Vector vector) -> org.apache.spark.mllib.linalg.Vectors.fromML(vector)
--- End diff --
This is an interesting point; what if the data is a mix of sparse and dense? At least this test now covers that case a bit.
The check above only tests the first element, but that's the only reasonable thing to do. If it were, for example, mostly sparse but the first happened to be dense, then this change could be problematic. I think it's OK to assume that most real-world uses are all sparse or dense. It is possible that some data in a sparse case is occasionally encoded as sparse as an optimization if it has a lot of zeroes, but this case works fine here.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237505113
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,69 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+ val dv = new DenseVector(v.toArray.zip(bc.value.toArray)
--- End diff --
I do some more research about your mention, the primitive array is more faster than zip, or zipped and view.zip. Because there is no more temporary collection and extra memory copies.
I do a comparison test, below is the result
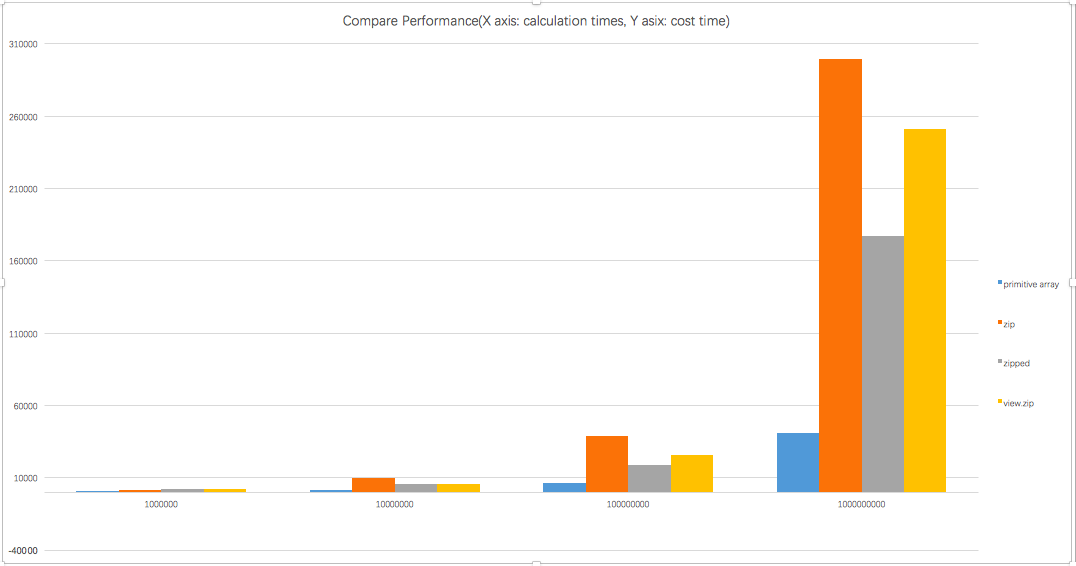
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237520303
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,82 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+
+ val n = v.size
+ val na = Array.ofDim[Double](n)
+ val means = bc.value
+ if (v.isInstanceOf[DenseVector]) {
+ v.foreachActive{(index, value) =>
--- End diff --
Nit (this might fail scalastyle): space around the braces here. But we don't need foreachActive here I think; they're all 'active' in a dense vector and every element needs the mean subtracted. Even for a sparse vector you have to do this. Do you really need a separate case here? we're assuming the vectors are (mostly) dense in this method.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237133038
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,69 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+ val dv = new DenseVector(v.toArray.zip(bc.value.toArray)
--- End diff --
A few more nits here. Let's broadcast `mean.toArray` to save converting it each time. Given this might be performance-sensitive, what about avoiding zip and computing the new array in a loop? I'm not sure how much difference it makes but I know we do similar things elsewhere.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
Um, the unit test in spark indeed cover both case. But there is function closeToZero to handle accuracy problem, so..
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
Add test case in RowMatrixSuite for this PR,
The breeze output is
6.711333870761802E-11 -3.833375461575691E-12
-3.833375461575691E-12 2.916662578525011E-12
Before add this commit, the result is:
1.9073486328125E-6 3.814697265625E-6
3.814697265625E-6 -7.62939453125E-6
After add this commit, the result is:
6.711333870761802E-11 -3.833375461575691E-12
-3.833375461575691E-12 2.916662578525011E-12
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237541497
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,82 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+
+ val n = v.size
+ val na = Array.ofDim[Double](n)
+ val means = bc.value
+ if (v.isInstanceOf[DenseVector]) {
+ v.foreachActive{(index, value) =>
--- End diff --
But isn't it incorrect to not subtract the mean from 0 elements in a sparse vector anyway?
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on the issue:
https://github.com/apache/spark/pull/23126
Is there a simple test case you can add to cover that too? that would really prove this change.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
Sure, the test cases include sparse and dense case.
Do these case again for new commit
we use data from
http://archive.ics.uci.edu/ml/datasets/EEG+Steady-State+Visual+Evoked+Potential+Signals
as **dense case**
we use data from
http://archive.ics.uci.edu/ml/datasets/Condition+monitoring+of+hydraulic+systems
as **sparse case**
Upload the test result
Dense data case
[sparkdensedataout.txt](https://github.com/apache/spark/files/2615937/sparkdensedataout.txt)
[numpydensedataout.txt](https://github.com/apache/spark/files/2615938/numpydensedataout.txt)
Sparse data case
[sparksparsedataout.txt](https://github.com/apache/spark/files/2615943/sparksparsedataout.txt)
[numpysparsedataout.txt](https://github.com/apache/spark/files/2615944/numpysparsedataout.txt)
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by srowen <gi...@git.apache.org>.
Github user srowen commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r236656961
--- Diff: mllib/src/test/scala/org/apache/spark/mllib/linalg/distributed/RowMatrixSuite.scala ---
@@ -266,6 +266,16 @@ class RowMatrixSuite extends SparkFunSuite with MLlibTestSparkContext {
}
}
+ test("dense vector covariance accuracy (SPARK-26158)") {
+ val rdd1 = sc.parallelize(Array(100000.000004, 100000.000012, 99999.9999931, 99999.9999977))
--- End diff --
I think you could just parallelize a couple DenseVectors, or (x,y) tuples and transform, rather than this way, but it doesn't matter really.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
Plug do some more test on real data after add this commit
we use data from
http://archive.ics.uci.edu/ml/datasets/EEG+Steady-State+Visual+Evoked+Potential+Signals
and data from
http://archive.ics.uci.edu/ml/datasets/Condition+monitoring+of+hydraulic+systems
to do some more accuracy test, the accuracy result is OK
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on a diff in the pull request:
https://github.com/apache/spark/pull/23126#discussion_r237551217
--- Diff: mllib/src/main/scala/org/apache/spark/mllib/linalg/distributed/RowMatrix.scala ---
@@ -128,6 +128,82 @@ class RowMatrix @Since("1.0.0") (
RowMatrix.triuToFull(n, GU.data)
}
+ private def computeDenseVectorCovariance(mean: Vector, n: Int, m: Long): Matrix = {
+
+ val bc = rows.context.broadcast(mean)
+
+ // Computes n*(n+1)/2, avoiding overflow in the multiplication.
+ // This succeeds when n <= 65535, which is checked above
+ val nt = if (n % 2 == 0) ((n / 2) * (n + 1)) else (n * ((n + 1) / 2))
+
+ val MU = rows.treeAggregate(new BDV[Double](nt))(
+ seqOp = (U, v) => {
+
+ val n = v.size
+ val na = Array.ofDim[Double](n)
+ val means = bc.value
+ if (v.isInstanceOf[DenseVector]) {
+ v.foreachActive{(index, value) =>
--- End diff --
Just, I do a quick test, input data
val data = Seq(
Vectors.dense(100000.000004, 199999.999999),
Vectors.dense(100000.000012, 0.0).toSparse,
Vectors.dense(99999.9999931, 200000.000003),
Vectors.dense(99999.9999977, 200000.000001)
)
and
val data = Seq(
Vectors.dense(100000.000004, 199999.999999),
Vectors.dense(100000.000012, 0.0),
Vectors.dense(99999.9999931, 200000.000003),
Vectors.dense(99999.9999977, 200000.000001)
)
for all dense vector case, the breeze and spark realization (substract the mean from 0) get the same result
6.711333870761802E-11 -0.6866668021258175
-0.6866668021258175 1.00000000001E10
but if change the one dense vector to sparse vector and not substract the mean from 0, the spark get the result
6.711333870761802E-11 -0.17166651863796398
-0.17166651863796398 2.500000000024999E9
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark pull request #23126: [SPARK-26158] [MLLIB] fix covariance accuracy pro...
Posted by asfgit <gi...@git.apache.org>.
Github user asfgit closed the pull request at:
https://github.com/apache/spark/pull/23126
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by SparkQA <gi...@git.apache.org>.
Github user SparkQA commented on the issue:
https://github.com/apache/spark/pull/23126
**[Test build #4445 has finished](https://amplab.cs.berkeley.edu/jenkins/job/NewSparkPullRequestBuilder/4445/testReport)** for PR 23126 at commit [`17117d7`](https://github.com/apache/spark/commit/17117d70ae0e26b5a99c6ea5a572cfbaf83080ab).
* This patch passes all tests.
* This patch merges cleanly.
* This patch adds no public classes.
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org
[GitHub] spark issue #23126: [SPARK-26158] [MLLIB] fix covariance accuracy problem fo...
Posted by KyleLi1985 <gi...@git.apache.org>.
Github user KyleLi1985 commented on the issue:
https://github.com/apache/spark/pull/23126
align JavaPCASuite expected data process behavior with PCA function fit
---
---------------------------------------------------------------------
To unsubscribe, e-mail: reviews-unsubscribe@spark.apache.org
For additional commands, e-mail: reviews-help@spark.apache.org